
Packaged or unpackaged: appearance and transport of extracellular noncoding RNAs in the plant apoplast
Here, we discuss the current status of the controversial debate on the localization, transport, molecular interactions, and activities of different types of extracellular noncoding RNAs as highlighted by a new publication (1) of the group of Roges Innes, Indiana University, on RNAs in the apoplastic intercellular washing fluid (IWF) of Arabidopsis thaliana.
Since the astonishingly successful development of new vaccines based on ribonucleic acid—better known as RNA—the compound has attracted a lot of attention. In the field of medicine, RNA vaccines are seen as promising drugs for treating serious diseases, such as certain types of cancer and neurodegenerative diseases, and thus receive special public attention. Only a minority is aware of the potentially effective use of RNA technology in agriculture and pest control, even though the alternative has been hotly debated among experts long before the Corona pandemic. Even without the current political disaster, it was clear that global crop yields would have to roughly double in the next 40 years to meet the increasing food and energy demands of the growing population—and this global goal must be achieved in the face of a changing climate and associated extreme weather (2).
Therefore, it is very clear that humanity also will face immense challenges in the area of agricultural production in the coming years. One of the few possible answers from research lies in the knowledge-based development of improved active ingredients that must be highly effective, selective and yet uncompromisingly environmentally compatible and biodiversity-friendly. Biologicals, i.e., natural substances and beneficial microorganisms that meet this requirement for modern agents, are increasingly found in nature today—RNA is one of them. As a highly effective yet very selective Biological, RNA could actually play an important role in crop protection and production (3-6). A fundamental discovery in 2016 was that cotton plants are naturally capable of producing small RNAs (sRNAs), in this case micro RNAs (miRNAs), that inhibit the growth of Verticillium dahliae, a fungal pathogen that infects plants and causes Verticillium wilt diseases on many crops (7). Two miRNAs, miR166 and miR159, were exported to the fungal hyphae to target two Verticillium dahliae genes encoding a Ca2+-dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15), respectively, both essential for fungal virulence. An earlier discovery had already shown that fungal pathogens also produce sRNA that interact with target genes in the plants and thus weaken the plants’ immune system (8). This bidirectional exchange of sRNA was termed cross-kingdom RNAi and requires key enzymes of the RNA interference (RNAi) pathway.
The agronomic relevance of dsRNA was demonstrated in 2006 when Huang et al. (9) reported reduced nematode infectivity when root-knot nematodes (RKN) fed on Arabidopsis thaliana expressing a 16D10 dsRNA targeting the 16D10 gene that encodes a conserved secretory root growth-stimulating RKN peptide involved in parasitism. That sRNA can indeed travel from a plant to an attacking pathogen was demonstrated in 2010 by Patrick Schweizer‘s group, and these authors coined the term host-induced gene silencing (HIGS) (10). Transient accumulation of dsRNA and antisense RNA with sequence similarity to fungal mRNA in plant cells reduced the development of the biotrophic fungus Blumeria graminis on barley and wheat leaves. For instance, silencing the fungal effector Avr10 resulted in reduced fungal development in the absence but not in presence of the corresponding resistance gene MLa10.
Since then, the expression of dsRNAs with complementary sequences to genes of pathogens and pests has been used to build resistance in plants to various species of viruses, bacteria, oomycetes, fungi, nematodes and insects (11). However, it is not clear how the sRNAs are transferred between plant and pathogen cells. Considering the literature on RNA transport in mammalian tissues, it is plausible to assume that these extracellular RNAs (exRNAs) must either be tightly associated with RNA-binding proteins or encapsulated in EVs to avoid degradation (12,13). However, whether EVs and/or RNA-binding proteins are required for RNA secretion or movement within the plant apoplast is still controversial (14,15). How plant RNA is taken up by microbial pathogens and pests and whether the uptake process can be improved is an unresolved question that is also critical for agricultural practice. The rapid uptake of topical RNA, resulting in a mechanism called spray-induced silencing (SIGS) (16,17), is a prerequisite for successful use of RNA in crop protection. Nematodes possess the protein SID-1 (Systemic RNA Interference Defective), which enables RNA transmembrane transport and the binding of double-stranded RNA (18). Moreover, some but by no means all insect groups have SID-like transporters, and it is added that dsRNA-degrading nucleases are widespread in insect groups, so that control of insets by topical RNA is concentrated in a few amenable groups (6,19). Fungi most likely do not have such channels. However, treatment of barley leaves with topical RNA resulted in its uptake by the leaf-colonizing Fusarium graminearum fungus (16); and leaves and fruits treated with topical RNA also showed reduced symptoms of the grey mould B. cinerea (17), suggesting that at least some fungi can take up RNA from the environment. Consistent with this, the uptake mechanism in Sclerotinia sclerotiorum occurs through clathrin-mediated endocytosis (CME). RNAi-mediated knockdown of several CME transcripts confirmed the involvement of this cellular uptake process in facilitating RNAi in S. sclerotiorum (20).
But what is the uptake mechanism for RNAs produced naturally in plants as cross-kingdom sRNAs or as products of a transgene (HIGS sRNAs)? A summary of the current state of knowledge is shown in Figure 1. In a landmark publication, it was reported that cells of Arabidopsis thaliana secrete exosome-like extracellular vesicles containing sRNA cargo that can subsequently be detected in the leaf-colonizing grey mold pathogen Botrytis cinerea (22). These sRNA-containing vesicles accumulate at infection sites and are taken up by fungal cells. A clear indication of sRNA uptake in this experiment was evidence of their RNAi activity, as they induced silencing of fungal target genes critical for pathogenicity. This EV-mediated RNA transport and delivery, although it seems plausible given the wealth of literature on mammalian EV cargo, is partially challenged by a recent publication by Roger Innes’ group (1). In their work they found less evidence of EV-mediated transport claiming that the majority of apoplastic RNAs are not bound to EVs but rather protected by RNA-binding proteins. The starting point of their work was previous findings that the intercellular wash fluid (IWF) of healthy (non-infected) Arabidopsis thaliana leaves contains different types of sRNAs, including miRNAs (21–22 nucleotides, nt), small interfering RNAs (siRNAs; 21–24 nt), and less defined tiny RNAs (tyRNAs; 10–17 nt) with unknown functions (23,24). Only the apoplastic tyRNAs copurified with EVs when using a density gradient, while siRNAs and miRNAs were largely missing from density gradient-purified EVs, although they were present in total IWF. The authors interpreted this then to mean that EVs may not be the primary carrier of apoplastic siRNAs and miRNAs (23). They further argued that in light of previous publications, it cannot be ruled out that large RNA-protein complexes copurify with EVs or that RNAs adhere to the surface of EVs, questioning that all RNAs are cargo of EVs. To eliminate extravesicular RNA-protein complexes and RNA adhering to the surface of EVs, most previous work used micrococcal nuclease treatment but refrained from treating purified EVs first with proteases to remove any RNA-binding proteins and then with nuclease to degrade unprotected RNAs. To distinguish EV-bound cargo RNA fractions from non-EV cargo RNA fractions, Zand Karimi et al. (1) now isolated IWF of Arabidopsis leaves by centrifugation (P40 fraction), and subsequently treated the pellet first with the protease trypsin and then RNase A, resulting in a strong shift of the portion of these RNAs vs. RNA isolated without treatment. Importantly, combined treatment reduced the amount of sRNA dramatically with only a few sRNA species resisting the treatment. A deeper differential expression analysis identified seven miRNAs inside EVs after trypsin and RNase treatment. Interestingly, six of the seven identified miRNAs corresponded to passenger strands of active miRNAs, raising the question if those miRNAs have a role in silencing. The same analysis was applied by focusing on apoplastic trans-acting (tasi)RNAs, including Tas1c_16_461 and Tas2_0_56, that are believed to be involved in cross-kingdom RNAi (22). Consistent with their reasoning, Zand Karimi et al. (1) were also unable to detect these tasiRNA after treatment of the apoplastic sample with trypsin and nuclease, so none of them appeared to be protected and enriched in the EVs. Although the above findings seem comprehensible, it must be pointed out that in the Zamt Karimi reference uninfected Arabidopsis plants were used as biological material; at least it cannot be completely excluded that RNA-loaded vesicles are secreted especially in response to biological stress. Moreover, despite the abundance of current literature, plant research on EVs and exRNA carriers is still very challenged and limited, especially by (I) the heterogeneity of EVs and their RNA cargo (differences between cell types and organs); (II) the sample-to-sample heterogeneity of EVs and other RNA carriers; (III) the differences in the methods used to isolate EVs and other RNA carriers; (IV the differences in sensitivity, specificity, reproducibility, and bias of different RNA profiling methods; and last but not least, the bias in the knowledge of plant EVs due to the predominantly mammalian-focused literature on EVs and their RNA carriers (12).
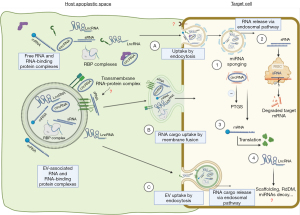
Of note, further analysis of apoplastic RNA detected both sRNAs and long noncoding RNAs (lncRNAs), including circular RNAs (circRNAs), and these RNAs were highly enriched in the posttranscriptional modification N6-methyladenine (m6A) (1). Consistent with this, the putative m6A-binding protein GLYCINE-RICH RNA-BINDING PROTEIN 7 (GRP7) as well as the sRNA-binding protein AGO2 was found in the IWF. These two proteins coimmunoprecipitated with lncRNAs, including circRNAs. Mutation of GRP7 or AGO2 caused changes in both the sRNA and lncRNA content of IWF, suggesting that these proteins contribute to the secretion and/or stabilization of apoplastic RNA. Previous work already identified already several RNA-binding proteins in the apoplast of Arabidopsis leaves, including AGO1, ANNEXIN1 and 2, and RNA HELICASE11 and 37 (15). However, the exact localization of these proteins, like that of RNA, is unclear, as definitive proof of their localization in EVs would require at least protease protection assays showing that these proteins remain undigested in the presence of trypsin and thus are located within EVs.
The circRNA, now also discovered in IWF from Arabidopsis leaves, is increasingly becoming the focus of research in plant pathology. In mammals, circRNAs were initially associated with their function as miRNA sponges that influence mRNA turnover (25). More recent findings show their function in regulating transcription and translation, sequestering and translocating proteins, facilitating interactions between proteins, and translating to proteins (26). Of note, the RNA base modification m6A, now found in IWF, is known to efficiently initiate translation of circRNAs (27). Cytoplasmic circRNAs can arise from exons (exonic circRNA) (28) and are thus the candidates for secretion into the apoplast. circRNA has also been detected in plants such as rice (Oryza sativa) and Arabidopsis thaliana (29) and they seem to accumulate in response to biotic stress (30) and abiotic stress, such as phosphate-sufficient and -starvation conditions. Due to the structural stability of the circular form and its potentially multifunctional roles they may play in plant pathogens and pests, circRNAs are promising candidates for agronomical application.
Problems to be solved: future application in crop protection
The use of RNA in agriculture has become a realistic scenario in recent years. Not surprisingly for scientists in this field, GMO approaches such as HIGS have proven to be particularly effective. What does the Zand Karimi paper (1) tell us about the future application of RNA in crop production? While the uptake of RNAi-inducing dsRNA by microbial pathogens and pests from the environment is uncontroversial, questions remain, such as how exactly RNA is transferred from the plant host to a colonizing microbe and what type of (apoplastic) RNAs play a key role in influencing the growth of a pathogenic microbe on its host.
What are the pitfalls that have to be avoided in order to introduce the RNA approach to the market? First of all, the products would need to be taken up more efficiently into the plants and pathogens/pests, even under adverse weather conditions, and the RNA must be protected – as it is under natural conditions by RNA-binding proteins and/or vesicles –over a certain period of time in order to enter the plants without being inactivated. What seems optimal for the environmental effect of RNA, namely its very rapid degradation in arable soil, especially by ubiquitous microorganisms and their RNA-degrading enzymes, the nucleases, is initially a disadvantage for the efficacy of applied RNA in crop protection. Therefore, the greatest challenge in RNA research is to package the RNA to improve its uptake by pathogens and pests. So, as with RNA vaccines, the biggest problem in crop protection is getting the very “sensitive” molecule to its site of action. Analogous to RNA vaccines, packaging the RNA in lipid droplets, for example, could also provide a solution. Moreover, the size and structure of the RNA molecule is critical for uptake by a pathogen and its RNAi activity (31). In some insects, it has been found that only RNA molecules with more than 60 nt are effective (6). In fungi, the optimal length of RNA molecules to be administered is not well understood. An emerging possibility is the use of circRNA, as this type of RNA is ubiquitous and seems to be more stable and possibly more efficient than dsRNA.
Acknowledgments
Funding: We thank the German Research Foundation (DFG) for supporting our research on RNA in plant-pathogen interactions by granting the DFG research unit RU5116 on exRNA. SN is supported by the Dr. Ernst-Leopold Klipstein-Stiftung, Paderborn, Giessen.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-11/coif). The research has been funded by the Dr. Ernst-Leopold Klipstein Foundation and a publication license was grant to Sabrine Nasfi for use in journal publications. The authors have no other conflict of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zand Karimi H, Baldrich P, Rutter BD, et al. Arabidopsis apoplastic fluid contains sRNA- and circular RNA-protein complexes that are located outside extracellular vesicles. Plant Cell 2022;34:1863-81. [Crossref] [PubMed]
- Brooks J, Deconinck K, Giner C. Three Key Challenges Facing Agriculture and How Start Solving Them. 2019. Accessed November 18, 2021. Available online: https://www.oecd.org/agriculture/key-challenges-agriculture-how-solve
- Cai Q, He B, Kogel KH, Jin H. Cross-kingdom RNA trafficking and environmental RNAi-nature's blueprint for modern crop protection strategies. Curr Opin Microbiol 2018;46:58-64. [Crossref] [PubMed]
- Taning CN, Arpaia S, Christiaens O, et al. RNA-based biocontrol compounds: current status and perspectives to reach the market. Pest Manag Sci 2020;76:841-5. [Crossref] [PubMed]
- Fletcher SJ, Reeves PT, Hoang BT, et al. A Perspective on RNAi-Based Biopesticides. Front Plant Sci 2020;11:51. [Crossref] [PubMed]
- Liu S, Jaouannet M, Dempsey DA, et al. RNA-based technologies for insect control in plant production. Biotechnol Adv 2020;39:107463. [Crossref] [PubMed]
- Zhang T, Zhao YL, Zhao JH, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2016;2:16153. [Crossref] [PubMed]
- Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013;342:118-23. [Crossref] [PubMed]
- Huang G, Allen R, Davis EL, et al. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A 2006;103:14302-6. [Crossref] [PubMed]
- Nowara D, Gay A, Lacomme C, et al. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010;22:3130-41. [Crossref] [PubMed]
- Koch A, Wassenegger M. Host-induced gene silencing - mechanisms and applications. New Phytol 2021;231:54-9. [Crossref] [PubMed]
- Zhang Y, Liu Y, Liu H, et al. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci 2019;9:19. [Crossref] [PubMed]
- O'Brien K, Breyne K, Ughetto S, et al. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol 2020;21:585-606. [Crossref] [PubMed]
- Rutter BD, Innes RW. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol 2017;173:728-41. [Crossref] [PubMed]
- He B, Cai Q, Qiao L, et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plants 2021;7:342-52. [Crossref] [PubMed]
- Koch A, Biedenkopf D, Furch A, et al. An RNAi-Based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog 2016;12:e1005901. [Crossref] [PubMed]
- Wang M, Weiberg A, Lin FM, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2016;2:16151. [Crossref] [PubMed]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 2002;295:2456-9. [Crossref] [PubMed]
- Cooper AM, Silver K, Zhang J, et al. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag Sci 2019;75:18-28. [Crossref] [PubMed]
- Wytinck N, Sullivan DS, Biggar KT, et al. Clathrin mediated endocytosis is involved in the uptake of exogenous double-stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum. Sci Rep 2020;10:12773. [Crossref] [PubMed]
- Joshi BS, de Beer MA, Giepmans BNG, et al. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020;14:4444-55. [Crossref] [PubMed]
- Cai Q, Qiao L, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018;360:1126-9. [Crossref] [PubMed]
- Baldrich P, Rutter BD, Karimi HZ, et al. Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019;31:315-24. [Crossref] [PubMed]
- Budak H, Kaya SB, Cagirici HB. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front Plant Sci 2020;11:276. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- He AT, Liu J, Li F, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther 2021;6:185. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Zhang P, Li S, Chen M. Characterization and Function of Circular RNAs in Plants. Front Mol Biosci 2020;7:91. [Crossref] [PubMed]
- Fan J, Quan W, Li GB, et al. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiol 2020;182:272-86. [Crossref] [PubMed]
- Šečić E, Kogel KH. Requirements for fungal uptake of dsRNA and gene silencing in RNAi-based crop protection strategies. Curr Opin Biotechnol 2021;70:136-42. [Crossref] [PubMed]
Cite this article as: Nasfi S, Kogel KH. Packaged or unpackaged: appearance and transport of extracellular noncoding RNAs in the plant apoplast. ExRNA 2022;4:13.


