
Targeting EGFR-steered cancer via erasing a circular RNA
Circular RNAs (circRNAs) are a type of long non-coding RNA (lncRNA), which form when a 5’ splice site binds to an upstream 3’ splice site and a circular structure is the result, also referred to as back splicing (1). Unlike other forms of RNA, circRNA is highly stable and conserved, due to its ability to resist the activity of RNA exonucleases, such as RNase R (2). CircRNAs can be localized in the nucleus, cytoplasm, and mitochondria. Those located in the nucleus are associated with genome editing, while those in the cytoplasm can act as miRNA sponges, as well as interacting with other proteins (3). A recent study has shown that downregulated steatohepatitis-associated circRNA ATP5B regulator (SCAR) is localized at mitochondria of liver fibroblasts from patients with nonalcoholic steatohepatitis (4). Decreased levels of circRNA SCAR switches on mitochondrial permeability transition pore by releasing ATP5B leading to increased mitochondrial ROS output. The interaction between circRNAs and miRNAs has been discussed in lung and other tumors in previous reviews (5,6). For example, circ_001569 has been shown to promote the proliferation of tumor cells in colorectal cancer through the sponging of miR-145, whose functional targets include BAG4, a regulator of apoptosis (7). There are many potential clinical applications of circRNAs. Currently, there are studies that suggest that it can be used as a biomarker, useful for detecting certain cancers, such as gastric cancer and oral squamous cell carcinoma (8,9). Currently there is a clinical trial, in the recruiting phase, that are exploring the reliability of circRNAs as biomarkers and potential diagnostic markers for pancreatic and biliary duct cancers (clinicaltrials.gov).
In a novel study conducted by Ishola et al. (10), in the Cancer Research journal, they introduce potential roles for circRNA, hsa_circ_0000190 (C190) in lung cancer. In a previous study, the authors discovered hsa_circ_0000190 and identified it as a biomarker for non-small cell lung cancer (NSCLC) due to its ease of detectability in patients' blood (11). One of the miRNAs that can be sponged by C190, is miR-142-5p, as validated by Ishola et al. (10). MiR-142-5p has been shown to play a role in the suppression of lung cancer through inhibition of cell migration and proliferation, as well as immune system modulation (12,13). The author’s main objective in this study was to identify the major functions of C190 in lung cancer progression. After analyzing transcriptomes of various lung cancer cell lines, they found that C190 had been the most upregulated circRNA. They confirmed expression of C190 in NSCLC patients’ blood samples using RNA-FISH and RT-qPCR techniques. Using the following cell lines: HCC827, A549, H1299 and BEAS-2B, they found that overexpression of C190 led to increased cell proliferation as well as increased phosphorylation of ERK1/2, not phosphorylation of epidermal growth factor receptor (p-EGFR), which may suggest that there is no direct positive or negative feedback loop with EGFR and C190. Additionally, the authors found that after conducting a wound-healing experiment, C190 increased cell migration. To test the effects of this circRNA on tumor growth, they stably transfected A549, H1299, and HCC827 cells with C190 and found that, through activation of the MAPK/ERK pathway, cell proliferation increased. Another mechanism of C190 is regulation of the cell cycle. By performing a transient transfection of the A549 cells, they found that cyclin-dependent kinase 1 (CDK1) and CDK4/6 are upregulated, while retinoblastoma (Rb) is hyperphosphorylated. This indicates that there is a positive correlation between upregulation of the cell cycle and EGFR activation through a C190-driven mechanism.
This study was conducted with a two-pronged perspective of both overexpression and knockdown of C190. This is advantageous because it confirms the function of the protein of interest in a holistic way, while also revealing potential significant protein-protein interactions. In this paper, for example, they found that when C190 was overexpressed in A549 and H1299, levels of phosphorylated ERK1/2 and ribosomal protein S6 (RPS6) were elevated. ERK1/2 is a downstream effector of EGFR and RPS6 is a component of the 40S ribosomal subunit and thus involved in protein translation, which further supports their hypothesis that C190 is linked to EGFR activation and modulation of global translational mechanisms. However, when C190 was knocked down using CRISPR-Cas13 RNA editing, the same proteins were downregulated in HCC827, a cell line with a constitutively active EGFR mutation. This suggests that there may be an alternative mechanism of C190 that is independent of EGFR activation or an alternative regulator of the MEK/ERK pathway and RPS6. The use of CRISPR-Cas13 RNA editing system allowed the authors to reduce off-target effects of C190 knockdown, which increases the reliability of their results. Furthermore, they demonstrated the clinical relevance of their work, by incorporating the function of the CRISPR-Cas13 editing system with the effects of EGFR-tyrosine kinase inhibitors (TKIs), as well as showing the pro-oncogenic effects of C190 in vivo using a mouse tumor model.
For future studies on C190, different cell lines with alternate mutations can be analyzed to see if the functions of C190 are restricted to the development of NSCLC or if it can play a role in other kinds of lung cancers. In Fig. 2M, for example, gefitinib treatment in KRAS mutant A549 (EGFR wild-type, non-responder to gefitinib) did not change C190 expression. EGFR mutant cell line HCC827 and other cell lines should be used to re-evaluate this effect. Another point of consideration is that there may be a time-dependent activation of EGFR that could affect downstream targets. At various times, the stimulation of EGF will lead to preferential phosphorylation of certain sites (14,15). That could be why STAT3 phosphorylation was not observed in H1299 and A549. In Fig. 6D, there may be concern about emergence of drug resistance after targeting C190 via the cancer cells utilizing other signaling pathways, such as the EGFR-phosphoinositide 3 kinase (PI3K) signaling pathway. Blocking C190 did not change the p-EGFR expression, which can induce other signaling pathways such as PI3K to bypass the C190-MAPK signaling pathway.
While this study brings to light many of the mysteries surrounding circRNAs and their functions in cancer progression, further exploration will be needed to understand the differences in functions of linear RNAs and circular RNAs that share the same promoter, for example CNIH4 and C190, as mentioned in the paper. Additionally, we want to understand the frequency of exon splicing that leads to the formation of circRNAs. The exact role of C190 seems to vary across different cancers as well. In some cancers, such as multiple myeloma and gastric cancer, C190 was downregulated and that led to cell proliferation through derepressing miR-767-5p and miR-1252-5p respectively, which are other known targets of C190 (16,17). Another target of C190 is miR-382-5p, which has been shown to function as a tumor suppressor in colorectal cancer through its inhibition of KLF12 and HIPK3 (11,18). For future clinical trials that could be based on this C190, we may need to consider these possible off-target effects (19) of C190’s alternative sponging of miR-767, miR-1252, and miR-382 (Figure 1).
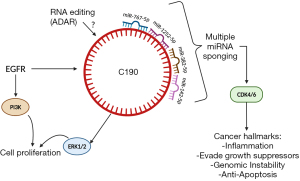
Since this paper showed that miR-142-5p, a guide strand of the miRNA duplex, and C190 have antagonistic roles, there should be further exploration into the function of miR-142-3p, a passenger strand, since it has also been shown to regulate the cell cycle (20). Also, the roles of miR-142-5p and C190 are suggested here to be time dependent, which could provide insight into clinical treatment efficacy. It is not clear if miR-142-5p will affect drug response to anticancer drugs, such as EGFR-TKIs. Several studies have shown that miRNAs mediate drug resistance such as EGFR-TKIs in lung cancer through different mechanisms (21-23). RNAs including miRNAs can be edited by an RNA editing enzyme such as adenosine deaminase RNA specific (ADAR) tool (24). Thus, how circRNA C190 and miR-142-5p are modified and edited by ADAR will be of interest in the future.
Acknowledgments
Funding: This study was supported by grants from the International Association for the Study of Lung Cancer Young Investigator Award, UCF Exploratory Research Award and UCF CBHRT (Center for Behavioral Health Research and Training) Award (to WCZ).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-9/coif). WCZ is supported by grants from the International Association for the Study of Lung Cancer Young Investigator Award, UCF Exploratory Research Award, UCF CBHRT (Center for Behavioral Health Research and Training) Award, and Atomwise Inc. AIMS Project A19-053. ARF has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015;5:472-80. [PubMed]
- Zhang J, Zhang X, Li C, et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol 2019;16:220-32. [Crossref] [PubMed]
- Zhao Q, Liu J, Deng H, et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020;183:76-93.e22. [Crossref] [PubMed]
- Santos RM, Moreno C, Zhang WC. Non-Coding RNAs in Lung Tumor Initiation and Progression. Int J Mol Sci 2020;21:2774. [Crossref] [PubMed]
- Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell 2019;179:1033-55. [Crossref] [PubMed]
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. [Crossref] [PubMed]
- Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 2017;466:167-71. [Crossref] [PubMed]
- Hung KF, Chen BH, Wang TY, et al. Identification of plasma hsa_circ_0000190 and 0001649 as biomarkers for predicting the recurrence and treatment response of patients with oral squamous cell carcinoma. J Chin Med Assoc 2022;85:431-7. [Crossref] [PubMed]
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res 2022;82:75-89. [Crossref] [PubMed]
- Luo YH, Yang YP, Chien CS, et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers (Basel) 2020;12:1740. [Crossref] [PubMed]
- Wang Z, Liu Z, Fang X, et al. MiR-142-5p Suppresses Tumorigenesis by Targeting PIK3CA in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2017;43:2505-15. [Crossref] [PubMed]
- Zhou C, Zhang Y, Yan R, et al. Exosome-derived miR-142-5p remodels lymphatic vessels and induces IDO to promote immune privilege in the tumour microenvironment. Cell Death Differ 2021;28:715-29. [Crossref] [PubMed]
- Begley MJ, Yun CH, Gewinner CA, et al. EGF-receptor specificity for phosphotyrosine-primed substrates provides signal integration with Src. Nat Struct Mol Biol 2015;22:983-90. [Crossref] [PubMed]
- Zheng Y, Zhang C, Croucher DR, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 2013;499:166-71. [Crossref] [PubMed]
- Feng Y, Zhang L, Wu J, et al. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res 2019;38:54. [Crossref] [PubMed]
- Wang GJ, Yu TY, Li YR, et al. Circ_0000190 suppresses gastric cancer progression potentially via inhibiting miR-1252/PAK3 pathway. Cancer Cell Int 2020;20:351. [Crossref] [PubMed]
- Yao H, Xia D, Li ZL, et al. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep 2019;39:BSR20180441. [Crossref] [PubMed]
- Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res 2011;28:2996-3015. [Crossref] [PubMed]
- Zhu X, Ma SP, Yang D, et al. miR-142-3p Suppresses Cell Growth by Targeting CDK4 in Colorectal Cancer. Cell Physiol Biochem 2018;51:1969-81. [Crossref] [PubMed]
- Zhang WC, Slack FJ. MicroRNA-21 Mediates Resistance to EGFR Tyrosine Kinase Inhibitors in Lung Cancer. J Thorac Oncol 2017;12:S1536. [Crossref]
- Zhang WC. microRNAs Tune Oxidative Stress in Cancer Therapeutic Tolerance and Resistance. Int J Mol Sci 2019;20:6094. [Crossref] [PubMed]
- Zhang WC, Wells JM, Chow KH, et al. miR-147b-mediated TCA cycle dysfunction and pseudohypoxia initiate drug tolerance to EGFR inhibitors in lung adenocarcinoma. Nat Metab 2019;1:460-74. [Crossref] [PubMed]
- Zhang WC, Slack FJ. ADARs Edit MicroRNAs to Promote Leukemic Stem Cell Activity. Cell Stem Cell 2016;19:141-2. [Crossref] [PubMed]
Cite this article as: Rodriguez-Fuguet A, Zhang WC. Targeting EGFR-steered cancer via erasing a circular RNA. ExRNA 2022;4:17.


