
Noncoding RNAs and modulation of the EGFR/ERK pathway by circRNA C190 in non-small cell lung cancer
Ishola et al. (1) begin to elucidate the mechanism of circular RNA (circRNA) C190 as a mediator of epidermal growth factor receptor (EGFR) signaling in non-small cell lung carcinoma (NSCLC). An earlier publication from the same group first identified elevated levels of C190 in the blood from patients with advanced lung cancer, but the functional involvement of C190 in the progression of lung cancer remained unclear (2). This current study uses NSCLC cell lines, innovative techniques, and bioinformatics to begin to uncover the role of C190 NSCLC progression.
Lung cancer remains one of the primary causes of cancer-related mortality worldwide with a 5-year survival rate of less than 15% (3,4). Lung cancer is a heterogeneous disease that is divided into two main subtypes, small cell lung carcinoma and NSCLC (3). NSCLC is responsible for about 85% of lung cancer cases with a majority of patients presenting advanced disease at diagnosis (4). The identification of druggable targets in patients, such as EGFR, ALK, PI3K/AKT/mTOR, Ras-MAPK, RET, MET, BRAF and NTRK/ROS1 have led to the development of targeted therapies (4). However, developing resistance to these targeted therapies, such as EGFR tyrosine kinase inhibitors (TKIs), is common and typically occurs within 10 to 16 months of treatment (5). For example, mutations within the kinase domain binding pocket of EGFR can lead to constitutive activation through a ligand independent mechanism (5). These activating mutations stimulate 3 major signaling cascades, the mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK), phosphatidylinositol 3-kinase (PI3K/AKT/mTOR) and interleukin 6 (IL6)/Janus Kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) contributing to cell survival, proliferation, invasion, and migration, as well as drug resistance (3,6). Better understanding the roles of downstream regulators of the EGFR pathway will help with eliminating acquired drug resistance potentially by being able to develop therapies that target the signaling pathway without directly targeting EGFR itself.
Recent advances in high throughput sequencing techniques and bioinformatic analyses have indicated that 80% of the genome harbors active biochemical function, while only ~2% of the genome actually accounts for protein-coding genes (7). It is becoming increasingly apparent that the non-protein coding portion of the genome contains critical regulators of gene expression and plays a large role in both normal human physiology and pathologic conditions. Non-coding RNAs (ncRNAs) include long non-coding RNA (lncRNA), microRNA (miRNA), circular RNA (circRNA), small interfering RNA (siRNA), PIWI-interacting RNAs (piRNA), transfer RNA (tRNA), ribosomal RNA (rRNA) along with many others. Dysregulation of these ncRNAs has increasingly been shown to be relevant in many disease states such as tumorigenesis, neurological, cardiovascular, and developmental disorders among several others. More specifically, lung cancer has been shown to be highly associated with dysregulation of expression of ncRNAs such as lncRNAs, miRNAs and more recently, circRNAs.
LncRNAs are the most abundant form of ncRNA in the mammalian non-coding transcriptome and are defined as transcripts of more than 200 nucleotides in length that are not translated into functional proteins (8). lncRNAs are involved in several processes including gene imprinting, cell/tissue differentiation, cell structure integrity, cell cycle, regulation of immune response and development of cancers. Because of their roles in regulating multiple signaling pathways that result in gene expression changes, dysregulation of lncRNAs has been implicated in cancer progression. They can be either oncogenic lncRNAs, which regulate tumor suppressor miRNA levels and/or activity by selectively sponging them (8) or tumor suppressive lncRNAs, which interact with tumor suppressor targets such as p53 and PTEN (9). For example, HOTAIR (HOX transcript antisense RNA), MALAT1 (metastasis associated lung adenocarcinoma transcript 1), and ANRIL (antisense noncoding RNA in the INK4 locus) are all found to have increased expression and support increased tumor progression, migration and metastasis in breast, bladder, colorectal, lung and gastric cancers, supporting their role as oncogenes (10). On the other hand, MEG3 (maternally expressed gene 3), lincRNA-p21, GAS5 (growth arrest specific transcript 5) and TUG1 (taurine-upregulated gene 1) all had decreased expression in various cancers supporting their tumor suppressive roles (10). lncRNAs have especially been implicated and extensively investigated in lung cancer both as single lncRNA in multiple tumors as well as a lncRNA expression signature in lung cancer.
miRNAs are highly conserved and the most well-studied group of ncRNA molecules with lengths ranging from 20–25 nucleotides. They mediate post-transcriptional gene silencing by controlling the translation of mRNA into proteins for more than 60% of protein-coding genes and are involved in regulating many processes including differentiation, survival, proliferation and development (11). Some miRNAs regulate specific individual targets, while others can function as master regulators of an entire pathway and mediate the expression of many genes simultaneously (11). The structure of miRNAs allows them to target several hundreds of transcripts making them very potent regulators of several processes. Aberrant expression of miRNAs can impact numerous signaling pathways and can result in cancer development, progression, and metastasis. Dysregulation of miRNAs in cancer can occur through either epigenetic changes (hypo/hypermethylation of CpG islands) or genetic alterations which ultimately affect the production of primary miRNA transcript, processing into mature miRNAs and interactions with mRNA targets (12). Specific miRNAs can act as either oncogenes or tumor suppressors depending on context, but generally their expression profile in human tumors is characterized by a deficiency miRNA production that results in global miRNA downregulation (13). They have also been proven to play a fundamental role in drug resistance and as biomarkers for early diagnosis especially in non-small cell lung cancer (14).
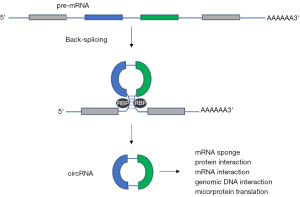
circRNAs are a more recently characterized group of ncRNAs that have covalently linked 3' and 5' ends. There are 3 different categories of circRNAs; exonic RNA (ecircRNA), exon-intron RNA (EIcircRNA) and intron RNA (ciRNA). The majority of circRNAs are ecircRNAs which are located in the cytoplasm, however EIcircRNA and ciRNA are typically located in the nucleus (15). Current studies suggest three models for the biogenesis of circRNA, direct back splicing, RNA-binding protein-mediated circularization, and lariat-driven circularization (15). They are the products of noncanonical pre-mRNA splicing, or backsplicing, that ultimately result in the formation of a closed loop RNA transcript (16) (Figure 1). This closed loop structure makes them more resistant to exonuclease digestion resulting in increased stability and half-lives up to 18.8–23.7 hours unlike their linear messenger RNA counterparts (15,17). Studies have shown that expression of circRNAs is developmentally regulated, tissue and cell-type specific, conserved across mammals and are regulated in a manner independent of their related linear isoforms (18). Because of their cell-type and organ specificity, it has been implicated that they have the potential to be used as biomarkers and therapeutic targets in the clinic (3,15). Similar to other noncoding RNAs, circRNAs play an important role in modulating transcription and translation (15). Depending on their sequence, circRNAs are involved in various biological functions including miRNA sponge activity, modulation of alternative splicing or transcription, interaction with RNA-binding proteins, sequestering and translocating proteins, facilitating protein interactions and others (15,19) (Figure 1). Dysregulated production, upregulation or downregulation of certain circRNAs has been implicated in several pathological conditions such as neurological disorders, diabetes, cardiovascular disease and importantly lung cancer development and in some instances confer drug resistance to small molecule inhibitors (20,21). Despite advances within the field, the role of circRNA in biological processes are still not well understood. Recent studies have begun to identify and elucidate the clinical utility and biological functions of specific isoforms, like circRNA_103625 and circRNA_0000190 (C190). Like C190, increases in circRNA_103625 expression in NSCLC patient samples is correlated to poor overall survival by Liang et al. (21).
Ishola et al. began their study by first confirming the upregulation of C190 in lung tumor tissue by performing RNA-FISH and digital droplet polymerase chain reaction (ddPCR) analysis on tumor biopsies and adjacent normal lung tissue from 21 NSCLC patients. While the C190 levels were increased in the tumor cells compared to the normal tissue, the levels of CNIH mRNA, the linear product of the C190-encoding gene, remained similar. Then, NSCLC cell lines were then used to elucidate whether the EGFR/MAPK pathway regulates C190 production and/or C190 regulates the MAPK/ERK pathway. When measured by quantitative polymerase chain reaction (qPCR), the H1299, A549, and HCC827 NSCLC cell lines were shown to produce C190 at estimated levels 5–20 times that of the normal bronchial epithelial cells, BEAS-2B. The authors rigorously verified that qPCR amplified the circRNA C190 and not the CNIH mRNA by (I) using primers that specifically only amplified the backsplice junction of C190 and comparing the successful amplification from cDNA rather than gDNA, (II) the qPCR products were subject to Sanger sequencing and were a perfect match to the backsplice junction of C190, and (III) showed the qPCR amplified C190 product from transcripts produced by A549 cells treated with actinomycin D had an increased stability, likely due to its circular structure, compared to the CNIH products.
Given the importance of the EGFR/MAPK pathway in lung cancer progression and having confirmed the ability to specifically assess relative amounts of C190, it was then determined that C190 was regulated by the MAPK/ERK signaling pathway. The A549 and H1299 cells were treated with EGF over time. Western blot analysis verified EGFR activation and downstream ERK1/2 and AKT activation within 15–30 min of EGF treatment in both cell lines, and qPCR revealed that C190 levels also began to increase over the same time. To determine whether the MAPK and/or PI3K pathway is responsible for C190 production, experiments were repeated in the presence of a MEK (PD98059) and PI3K (LY294002) inhibitor. Only the MEK inhibitor prevented the increase in C190 production after EGF treatment, and it is concluded that MEK activation is needed for C190 production. To determine whether EGFR activation upstream of MEK is required, gefitinib was used to block the EGFR kinase domain. It was concluded that the blockage of EGFR activation blocked EGF-induced activation of C190. However, the results in Fig. 2M are inconsistent with other data. Here, C190 production is greatly amplified after 15 min of EGF treatment, but instead of remaining high or increasing levels by 30 min, the levels drop significantly. If the C190 levels have already dropped by 30 min and the addition of gefitinib doesn’t change the level of C190 at 30 min, how can it be concluded that gefitinib was effective?
The next major conclusion from this work is that C190 can regulate the MAPK pathway to promote cell proliferation and migration. Both overexpression and targeted knockdown approaches are employed. Western blot analysis confirms the NSCLC cells transiently and/or stably overexpressing C190 also express elevated levels of activated ERK1/2, while the knock down of C190 expression reduces ERK1/2 activation. To reduce off target effects, a CRISPR/Cas13a editing system is used to target C190 more specifically and increase its degradation via Cas13a RNA endonuclease activity. Proliferation assays using transient overexpression of C190 in the A549 and HCC827 cells show that C190 can promote cell growth, while knockdown of C190 is the same cells reduces proliferation. Also, stable overexpression of C190 in A549 and H1299 cells promoted cell migration, while stable knockdown of C190 in the A549 cells impeded migration. However, it may be inaccurately concluded that C190 knockdown reduces anchorage free survival as it is unclear whether the cells were incubated in suspension for any period of time before plating in the clonogenic colony assay.
Since C190 could regulate in vitro proliferation, in vivo experiments were performed to determine if C190 could regulate tumor growth. C190 overexpression significantly increased tumor mass and Ki67 staining, while C190 knockdown inhibited tumor mass in only one of the crRNA transfected lines, even though both cRNAs equally reduced C190 expression and Ki67 staining. Additionally, some of the growth trends are not significant which may be due to a small sample size. Thus, the in vivo experiments elicit more questions, and more work will be needed to definitively determine whether C190 regulates NSCLC tumor growth.
Additionally, RNAseq comparative analysis was performed on the control A549 cells and their C190 overexpression counterparts. RNAseq and Expression-to-Kinase Network Analysis determined that C190 overexpression activates cell cycle and global translation kinases. Examination via western blot analysis confirmed the relative increase in Cdk1, Cdk4, and Cdk6 expression in the C190 overexpressing cells. To provide relevance and importance of the increase of Cdks in NSCLC patients, the authors searched the The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) database and showed that patients with high expression of Cdk1 or Cdk6 have significantly shorter survival times.
Finally, the authors determine that C190 indirectly targets Cdks, specifically Cdk6, via sponging miR-142-5p. TargetScan and CircInteractome databases were used to identify miR-142-5p as a target for both Cdk4 and Cdk6. Using RNA pulldown assays, qPCR, and luciferase reporter assays, C190 could directly bind miR-145-5p, miR-142-5p could bind the 3’UTR of Cdk6 to repress luciferase production, and upon C190 addition to miR-142-5p and the 3’UTR of Cdk6, luciferase repression was relieved. These results demonstrated that C190 can interrupt the binding of miR-142-5p to Cdk6 mRNA to allow for possible Cdk6 translation.
Since the discovery of circRNAs in the 1970’s, studies elucidating circRNA functions have been limited. But as more physiological and pathophysiological functions of circRNAs are discovered, there is an attempt to use circRNAs as biomarkers for cancer diagnosis, prognosis and/or to monitor drug response. Due to their circular structure, circulating circRNA are more advantageous than linear RNAs for non-invasive liquid biopsies. However, additional research is needed to reveal detailed mechanisms through which circRNAs regulate cancer progression.
Ishola et al. have begun to elucidate the mechanism of the circRNA, C190, in NSCLC. They confirmed C190 to be an oncogenic circRNA implicated in the progression of NSCLC and that C190 was both regulated by and exerted a positive effect on the activation of the MAPK pathway. Given the direct involvement in the EGFR/MAPK pathway downstream of EGFR activation, C190 may be an important target as a biomarker and/or for therapeutics to treat NSCLC patients.
Acknowledgments
Funding: American Cancer Society Research Scholar Grant No. 132153-RSG-18-028-01-CSM.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-12/coif). MIV is co-investigator on an NCI grant in which Stuart Martin is the PI. MIV is the PI on an American Cancer Society Research Scholars grant which will end 12/31/22. The funding is to examine cytoskeletal regulation in breast cancer cells during metastasis and never once mentions anything about nCrna. This first grant focuses on post transcriptional modifications of microtubules in unique membrane protrusions called microtentacles. The ACS grant is to examine the role of PTEN on actin signaling and cell deformation. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res 2022;82:75-89. [Crossref] [PubMed]
- Luo YH, Yang YP, Chien CS, et al. Plasma Level of Circular RNA has_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers (Basel) 2020;12:1740. [Crossref] [PubMed]
- Ishola AA, La’ah AS, Le HD, et al. Non-coding RNA and lung cancer progression. J Chin Med Assoc 2020;83:8-14. [Crossref] [PubMed]
- Majeed U, Manochakian R, Zhao Y, et al. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol 2021;14:108. [Crossref] [PubMed]
- Red Brewer M, Yun CH, Lai D, et al. Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc Natl Acad Sci U S A 2013;110:E3595-604. [Crossref] [PubMed]
- Ferreira RB, Law ME, Jahn SC, et al. Novel agents that downregulate EGFR, HER2, and HER3 in parallel. Oncotarget 2015;6:10445-59. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Yu JE, Ju JA, Musacchio N, et al. Long Noncoding RNA DANCR Activates Wnt/β-Catenin Signaling through MiR-216a Inhibition in Non-Small Cell Lung Cancer. Biomolecules 2020;10:1646. [Crossref] [PubMed]
- Ghafouri-Fard S, Shoorei H, Branicki W, et al. Non-coding RNA profile in lung cancer. Exp Mol Pathol 2020;114:104411. [Crossref] [PubMed]
- Khandelwal A, Bacolla A, Vasquez KM, et al. Long non-coding RNA: A new paradigm for lung cancer. Mol Carcinog 2015;54:1235-51. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Romano G, Veneziano D, Acunzo M, et al. Small non-coding RNA and cancer. Carcinogenesis 2017;38:485-91. [Crossref] [PubMed]
- He AT, Liu J, Li F, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther 2021;6:185. [Crossref] [PubMed]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015;4:e07540. [Crossref] [PubMed]
- Enuka Y, Lauriola M, Feldman ME, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 2016;44:1370-83. [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Xu S, Zhou L, Ponnusamy M, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ 2018;6:e5503. [Crossref] [PubMed]
- Chen T, Luo J, Gu Y, et al. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer 2019;10:930-41. [Crossref] [PubMed]
- Liang H, Lin Z, Lin H, et al. circRNA_103615 contributes to tumor progression and cisplatin resistance in NSCLC by regulating ABCB1. Exp Ther Med 2021;22:934. [Crossref] [PubMed]
Cite this article as: Chang KT, Ju JA, Vitolo MI. Noncoding RNAs and modulation of the EGFR/ERK pathway by circRNA C190 in non-small cell lung cancer. ExRNA 2022;4:21.


