
Adipogenesis-related microRNAs in obesity
Introduction
In the last four decades, studies have shown that the significant increase in the prevalence of obesity has become a pandemic. It is expected that by 2030, in the USA up to 86% of adults will be overweight or obese. Conspicuously, the proportion of the Australian population who are overweight and obese were reached to 66% in 2017. All-cause mortality rates in obese individuals and obesity incidence in different countries and communities are almost similarly high. While obesity decreases life expectancy approximately by 3.3 to 18.7 years, it largely increases healthcare expenditures (1,2). Compared with the metabolically healthy normal-weight individuals, obese persons are at increased risk for adverse long-term outcomes even in the absence of obesity-related diseases (3). In this context, up to 30% of obese patients are metabolically healthy like the healthy normal weight individuals (4). In unhealthy obese subjects, obesity-related diseases are associated with subcutaneous adipose tissue remodeling characterized by adipocyte hypertrophy, as well as chronic inflammation, hypoxia, increased visceral adipose tissue, and fatty liver. In contrast metabolically healthy obese individuals have a lower degree of ectopic fat accumulation, and visceral adipose tissue inflammation (5). Still, these obese individuals are at higher risk for cardiovascular diseases and type 2 diabetes compared with metabolically healthy normal weight individuals (4). Epidemiological data have shown that obesity is an important health problem worldwide and, in any case, it is characterized by an excessive increase in adipocyte hypertrophy and hyperplasia, as well as the inability to store excess fat into the lipid droplets (2,6). A pool of resident multipotent progenitor cells, which persists in adipose tissue throughout the life, differentiate to adipocytes and the adipocyte number increases. Because of hypertrophy and hyperplasia of adipocytes, adipose tissue enlarges. Thus, in widened fat mass in adipose tissue of obese individuals, both adipocyte size and numbers increase (7-10). Even after obese individuals undergo significant weight loss and regain a healthy appearance, elevated adipocyte number is maintained. In this context, adipogenesis, which is a permanent health problem in obese individuals, takes place in two stages; firstly, mesenchymal stem cells transform into preadipocytes. At second step, adipogenic stimuli induce terminal differentiation of these preadipocytes through the epigenomic activation of transcription factors (11). Transcription factors such as cytosine-cytosine-adenosine-adenosine-thymidine (CCAAT)/enhancer-binding proteins (C/EBPs) and peroxisome proliferator-activated receptor-gamma (PPARγ) play critical roles in the key events controlling the terminal differentiation of progenitor cells into adipocytes (12). Nonetheless, the mechanisms controlling the early steps of adipocyte progenitor cells differentiation to the adipocyte and molecular events regulating adipogenesis remain poorly understood. It is expected that functional analysis of adipocyte-derived microRNAs (miRNAs) can clarify the events that result in adipogenesis. Indeed, adipocyte-derived miRNAs have dual function in adipogenesis; while some of them stimulate the transcription factors that modulate adipocyte proliferation and differentiation, others block expression of master regulators of adipogenesis (13).
miRNAs are a subclass of regulatory, non-coding RNAs that properly adjust gene expression at a post-transcriptional level by affecting mRNA translation and stability (14). Up to 30% of human genes could potentially be regulated by miRNAs (15). In this context, the miRNAs involved in adipogenesis, are also part of the epigenetic mechanisms that regulate gene expression at a post-transcriptional level even in healthy obese individuals. The ability of a miRNA to share the same target while having diverse functions, constitute an increase in the complexity of the biological networks. The purpose of this review is to analyze the complex role of miRNAs in the regulation of adipogenesis.
Adipocyte-derived extracellular vesicles (AdEVs)
Induction of adipocyte stress and subsequent hypertrophy leads to the release of AdEVs that contain a unique cargo. As AdEVs release is significantly increased in hypertrophic adipocytes, properties, and the levels of circulating AdEV are accepted to be an important method to assess adipose tissue health. Therefore, proteomic analysis and miRNA sequencing of the cargo content of AdEV enables the identification of adipocyte-specific proteins and miRNAs that can be used as novel biomarkers (16,17). In fact, the cargo molecules of AdEVs are important messengers for intercellular communication involved in metabolic responses and have very specific signatures that direct the metabolic activity of target cells (18).
AdEVs involved in the exchange of components between donor and recipient cells are classified as microvesicles and exosomes, although their sizes are not always different (19). Both are generally employed as vehicles to transfer miRNAs (20,21). However, the contents of adipocyte-derived exosomes (AdEXs) mediate a diverse series of cellular responses, including adipogenic differentiation, also via other cargo molecules besides miRNA. Thus, in response to adipogenic stimuli, the endogenous lysosomal Ca2+ permeable channel transient receptor potential mucolipin 1 (TRPML1) expression in pre-adipocytes is increased in a time-dependent manner. Since the deletion of TRPML1 reduces expression of differentiation-related genes, particularly, mature AdEXs are necessary for adipogenesis (22). Thus, preadipogenic AdEX concentration is elevated prior to adipogenesis. Parent cells contain higher proportions of phosphatidylserine (PS) and show higher annexin V binding in comparison to recipient cell (23). These exosomes carry high levels of signaling fatty acids such as arachidonic acid, and the markers of adipogenesis such as PPARγ1 (Figure 1), as well as preadipocyte factor-1 (PREF1), which represses differentiation (23,24). Angiogenesis is expected to adapt to adipogenesis during the expansion of adipose tissue (25). However, in morbid obesity, angiogenesis is insufficient to achieve an appropriate level of vascularization for the expanded adipose tissue (26). Compared with lean subjects, adipose tissues of overweight/obese subjects have 44% lower capillary density and 58% lower vascular endothelial growth factor (VEGF). This is due to lower PPARγ1, and higher collagen VI mRNA expression, which correlates with the lower adipose tissue oxygen partial pressure (27). Although a total of 231 proteins have been identified in the AdEX, the total amount of proteins secreted from exosomes increases by three-four-fold under hypoxic conditions (28). It is known that hypoxia inhibits adipocyte differentiation from preadipocytes. Since the hypoxic AdEXs are enriched with enzymes related to de novo lipogenesis such as acetyl-CoA carboxylase, glucose-6-phosphate dehydrogenase, and fatty acid synthase (FASN), despite hypoxia, exosomal proteins derived from adipocytes can trigger lipogenic activity in neighboring preadipocytes (28,29). Obviously, miRNAs in AdEXs play a more important role in the circulation than the other cargo proteins. Therefore, this comprehensive review is focused on miRNA content of AdEXs. In fact, genes for adipocyte-derived exosomal miRNA are transcribed to a primary miRNA (pri-miRNA). The pri-miRNA is processed within the nucleus of adipocytes to a precursor miRNA (pre-miRNA) by Drosha, a class 2 Rnase III enzyme. The transport of pre-miRNAs to the cytoplasm is mediated by exportin-5. In the cytoplasm, pre-miRNAs are converted to mature miRNAs by Dicer Rnase III type protein (30). The generation of mature miRNAs critically depends on the Rnase-III enzyme, Dicer (31). Dicer coordinately regulates the inflammatory response and lipid metabolism in macrophages through enhancing fatty acid-fueled mitochondrial respiration. miRNA-10a promotes fatty acid oxidation, which mediated the lipolytic and anti-inflammatory effect of Dicer (32). The neuronal-deletion of the miRNA-processing enzyme Dicer leads to the development of rapid and transient obesity (33). Thus, deletion of Dicer in anorexigenic pro-opiomelanocortin-expressing cells results in obesity, characterized by hyperphagia, increased adiposity, hyperleptinemia, defective glucose metabolism and alterations in the pituitary-adrenal axis (34). Furthermore, loss of miRNA-103 and inactivation of Dicer gene in the adult neurons leads to chronic overactivation of the phosphatidylinositol-3-kinase (PI3K)-Akt-mammalian target of rapamycin (mTOR) pathway and severe hyperphagic obesity. Although the Dicer genes are deleted, the continuous stimulation of PI3K-Akt-mTOR pathway components by oligonucleotides mimicking miRNAs attenuates adiposity (35).
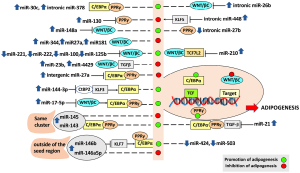
miRNA expression patterns in adipogenesis in animal and cell-line models
miRNAs profiles of adipocyte differentiation phase and of persistence obesity phase are quite different. The expression pattern of miRNAs during 3T3-L1 cell adipogenesis using miRNA microarrays detecting 373 mouse mature miRNAs have been analyzed. Among the 41 miRNAs upregulated during the adipocyte differentiation, 31 of the miRNAs are decreased during obesity. Contrarily, among the 38 miRNAs downregulated during the adipocyte differentiation, 26 miRNAs are increased in diet-induced obese mice. Although miRNA-103 and miRNA-143 are found to be important for adipogenesis, the specific mRNA targets of these miRNAs could not be detected (36). Dysfunctional adipocytes dysregulate the assembly and sequence of biological components in exosomes, leading to obesity-related diseases. Increase in palmitate-induced hypertrophic 3T3-L1 adipocytes-derived exosomal miRNA-802-5p content result in various alterations in the metabolic pathways, by downregulating the heat shock protein 60 (HSP60), increasing the expression levels of C/EBP-homologous protein. Enhanced oxidative stress, phosphorylation of c-Jun NH(2)-terminal kinase (JNK) and insulin receptor substrate-1 (IRS1), eventuate to the development of insulin resistance (37). Interestingly, the exosomal miRNAs secreted from the hypertrophic adipocytes are transferred to small adipocytes in the adipose tissues to promote lipogenesis and adipocyte hypertrophy by mediating horizontal transfer of lipogenic information. This lipogenic signal is encoded by relevant miRNAs and glycosylphosphatidylinositol (GPI)-anchored protein species. Thereby positive feedback on hypertrophic growth of rat adipocytes is transferred in a paracrine manner (38). During adipogenesis miRNAs not only can accelerate or inhibit adipocyte differentiation but also by regulating the adipogenic lineage commitment in multipotent stem cells, govern fat cell numbers (39).
Many miRNAs involved signaling pathways have differential effects on adipogenesis. Some miRNAs are employed to be negative regulators of adipocytes differentiation while others are capable of accelerating adipocyte differentiation (10). Therefore, co-transportation of miRNAs that activate or inhibit any of these signaling pathways within the same exosome elicits a complex effect on adipogenesis (40). Among the miRNAs whose expression levels change during differentiation of adipocyte, miRNA-146b dramatically increases by nine-fold and this miRNA promotes the adipogenesis in 3T3-L1 cells via suppression of sirtuin 1 (SIRT1) (41). SIRT1 catalyzes NAD+-dependent protein deacetylation and hinders adipogenesis by inhibiting PPARγ and stimulating lipolysis (42,43). The miRNA-146 family of miRNAs consists of two members, miRNA-146a and miRNA-146b. These two miRNAs are located on different chromosomes and conduct different functions in the body. Since the regulatory functions of miRNA-146a-5p and miRNA-146b are created by the sequence outside of the seed region, these miRNAs target different genes although they share the same seed region (44). miRNA-146a-5p suppresses adipogenesis (45). Whereas miRNA-146b inhibits the proliferation of visceral preadipocytes, while promoting their differentiation into mature adipocytes. Its binding site is in the 3' untranslated regions (3'UTR) of the Krüppel-like factor 7 (KLF7) gene, therefore miR-146b is a negative regulator of KLF7. In contrast, KLF5 is a transcription factor that activates the PPARγ2 promoter and regulates adipocyte differentiation (46). Ectopic expression of intergenic miRNA-27a in 3T3-L1 pre-adipocytes represses adipocyte differentiation by reducing PPARγ expression. In mature adipocyte fraction of obese mice, during the adipogenesis, the expression level of miRNA-27a is inversely correlated with that of adipogenic marker genes such as PPARγ and adiponectin. miRNA-27a suppresses adipocyte differentiation through targeting PPARγ. Contrarily, down-regulation of miRNA-27a is associated with adipose tissue enlargement in obesity (47). During adipocyte differentiation, changes in the miRNA profile of circulating AdEX precisely depend on obesity. Wnt/β-catenin signaling is a negative regulator of adipogenesis, and Wnt1 expression decreases significantly during the progression of adipocyte differentiation (48). In this context, there are 18 miRNAs that might promote adipogenesis by repressing Wnt signaling, such as miRNA-210, miRNA-148a, miRNA-194, miRNA-322. In contrast, 29 miRNAs show a negative effect on adipogenesis by activating Wnt signaling in 3T3-L1 cells, such as miRNA-344, miRNA-27 and miRNA-181 (49) (Figure 1). Following the induction of differentiation, increasing in the expression of miR-143 together with the miRNA-17 and miRNA-21 enhance the adipogenic differentiation in 3T3-L1 pre-adipocytes, and in porcine bone marrow-derived mesenchymal stem cells as in human preadipocytes. In all samples miRNAs stimulate the expression of adipogenic transcription factor, C/EBPα mRNA (50-52). Although miRNA-145 is classified into the same cluster with miRNA-143, miRNA-145 is a potent inhibitor of adipogenesis that may function by reducing the activity of PI3K/Akt and mitogen-activated protein kinase (MAPK) signaling pathways (53). Like miRNA-143, miRNA-103 is also abundantly produced by adipocytes of obese animals. Expression of miRNA-103 is induced approximately nine-fold during adipogenesis. Consequently, expression of adipogenic transcription factor, PPARγ2 is doubled by the expression of either miRNA-103 or miRNA-143. miRNA-103 increases the expression of fatty acid binding protein 4 (FABP4) and adiponectin approximately nine-fold and four-fold, respectively. FABP4 coordinates lipid transport in mature adipocyte and its inhibition in obesity results in weight loss and restores insulin sensitivity. Initially, upregulation of miRNA-103 and miRNA-143 during adipogenesis and downregulation after permanency of obesity suggest that persistence of obesity leads to miRNA loss. This finding is a sign that the mature adipose tissues of leptin deficient ob/ob, and diet-induced obese mice consist of fully differentiated and metabolically active adipocytes during adipogenesis (36,54). As with miRNA-143, miRNA-17 and miRNA-21, miRNA-375 expression is increased after induction of adipogenic differentiation. Overexpression of miRNA-375 accelerates adipocyte differentiation, via increasing mRNA levels of both C/EBPα and PPARγ2. Adipocyte FABP and triglyceride accumulation is simultaneously induced. The overexpression of miRNA-375 promotes the adipogenesis via 3T3-L1 adipocyte differentiation by suppressing phosphorylation of extracellular signal-regulated protein kinase (ERK1/2), which has opposing effect during adipogenesis (55). In the early phase of adipocyte differentiation, ERK is activated for a proliferative step, but later in preadipocyte differentiation, ERK needs to be shut off to prevent PPARγ phosphorylation (56). It is thought that increased expression of PPARγ2 and FABP is due to the inhibitory effect of ERK on PPARγ2. In this process, ERK activation is suppressed by miRNA-375 (55).
It is thought that in adipose tissue, forkhead box protein O1 (FOXO1) inhibits adipocyte differentiation by suppressing lipogenesis through PPARγ, which is a prerequisite for adipocyte differentiation. This anti-lipogenic effect of FOXO1 is controlled by insulin signals (57-59). miRNA-144 targets FOXO1, thus reducing its expression and inhibiting its promotional effect on adiponectin, thereby alleviating the inhibitory effect of adiponectin on adipogenesis in porcine pre-adipocytes (60). On the other hand, miRNA-146b facilitates adipocyte differentiation through the inhibition of SIRT1/FOXO1 cascade. The miRNA-146b/SIRT1 axis mediates adipogenesis through increased acetylation of FOXO1 in 3T3-L1 cells (41,61). It is claimed that both transcriptional and translational levels of several novel lipogenic genes are upregulated by overexpression of miRNA-15a/ba in the preadipocytes. Thereby, adipogenesis are promoted in pre-adipocyte via suppression of FOXO1. Although miRNA-15a/b expression that suppresses FoxO1 is relatively high in the early phase of adipocyte differentiation in porcine pre-adipocytes, it decreases later due to the dynamic change in FOXO1 expression (62).
AdEXs interact with the adipose tissue-derived stem cells (ADSCs) and macrophages to maintain the balance of metabolism and immunity. Consequently, they drive a vicious cycle creating immunometabolic imbalance between hypertrophic adipocytes and inflammatory macrophages (63). 3T3-L1 adipocytes’ AdEXs contain approximately 7,000 mRNAs and 140 miRNAs and mediate transport of adipokine gene transcripts into macrophages (64). Hypertrophic adipocyte-derived chemotactic monocyte chemoattractant protein-1 (MCP-1)/C-C chemokine receptor 2 (CCR2) pathway and hypoxia along with higher concentrations of free fatty acids are the main mechanisms for adipose tissue macrophage (ATM) accumulation and macrophage-mediated inflammation in obesity. Consequently, the ratio of M1-to-M2 macrophages is increased in obesity (65). Obesity induces imbalance in the M1-to-M2 macrophage ratio in adipose tissue. Adipocyte-derived exosomal miRNAs from obese mice induce M1 macrophage polarization (66). M1 polarization of ATMs is characterized by production of pro-inflammatory cytokines, whereas M2 polarized macrophages is linked to immunosuppression. miRNA-9, miRNA-127, miRNA-155, and miRNA-125b have been shown to promote M1 polarization while miRNA-124, miRNA-223, let-7c, miRNA-132, miRNA-146a, and miRNA-125a-5p induce M2 polarization in macrophages by targeting various transcription factors and adaptor proteins (67). Like obese mice, in obese patients, the elevated level of miRNA-155 in the AdEXs also induces pro-inflammatory M1-macrophage phenotype by activating signal transducer and activator of transcription 1 (STAT1) and repressing STAT6 expression (66). Contrarily, over-expressed miRNA-223 induces M2 macrophages polarization in mouse (68). Furthermore, transported miRNA-34a with the AdEXs from visceral adipocytes into macrophages suppresses M2 macrophage polarization by repressing KLF4, while inducing obesity-related adipose tissue inflammation in obese mice (69). Co-culture of adipocyte stem cells with ATMs of mice promote brown adipocyte gene expression upon differentiation. miRNAs derived from ATMs are transferred into adipocyte stem cells to facilitate beige adipogenesis, via exosomal transfer. Overexpression of ATM-derived miRNA-10a-5p effectively suppresses pro-inflammatory polarization of ATMs and promotes brown/beige differentiation of adipocyte precursor cells of mice. The ability to reduce levels of crown-like structures while promoting adipogenesis indicates that miRNA-10a-5p enhances tissue restoration and supports healthy adipose tissue remodeling (70).
De novo lipid biosynthesis is controlled by sterol regulatory element-binding proteins (SREBPs), which are also an important regulator of the PPARγ gene and promote the expression of important adipocyte-specific mRNAs 3T3-L1 cells (71). Some miRNAs, which are critical mediators in lipid metabolism and adipogenesis, are located in the SREBP-1 or SREBP-2 gene in humans (72). The primary transcript of SREBP2 contains an intronic miRNA-33 that increases lipid synthesis and prevents export and degradation of newly synthesized lipids in HepG2 cells (73). miRNA-33b is induced, along with SREBP-1, during differentiation of human preadipocytes. Overexpression of miRNA-33b causes reduced preadipocyte proliferation and impaired differentiation, while inhibition of miRNA-33b enhances lipid accumulation in differentiating adipocytes (74). If miRNA-33 inhibits the expression of cyclin-dependent kinase 6 (CDK6) and cyclin D1 (CCND1), cell proliferation and cell cycle progression are reduced after partial hepatectomy in mice (75). The chromatin-remodeling factor, high-mobility group AT-hook 2 (HMGA2), is upregulated during the clonal-expansion phase of adipogenesis but downregulated following terminal differentiation of 3T3-L1 cells (76). HMGA2, which has 3'-UTR, contains three predicted binding sites of miRNA-33a (77). Overexpression of miRNA-33b decreases both the protein and mRNA levels of HMGA2 in mouse. HMGA2 deficiency impairs adipocyte differentiation and the induction of PPARγ target genes in a manner like that observed with miRNA-33b overexpression (74). On the other hand, HMGA2, as a transcription factor that regulates the growth and proliferation, is inversely correlated with let-7 levels during adipogenesis (76).
The C/EBPα is an important transcription factor and regulator of adipogenesis involved in promoting the activation of PPARγ and its downstream targets. Immortalized line of fibroblasts lacking PPARγ have no ability to promote adipogenesis (78). As expression of C/EBPα is downregulated both prior to and after adipocyte differentiation, targeting of C/EBPα by miRNA-33b may play an important role in mediating the effects of miRNA-33b on adipogenesis and the induction of PPARγ targets (74). miRNA-20a-5p is a positive regulator of adipocyte differentiation and lipogenesis. C/EBPα transcriptionally activates miRNA-20a-5p expression via binding to the promoter of miRNA-20a-5p and induces adipocyte differentiation from 3T3-L1 preadipocytes (79). Overexpression of miRNA-144-3p suppresses the expression of cell cycle regulatory factors and inhibits pre-adipocytes proliferation in obese mice. Besides, overexpression of miRNA-144-3p accelerates lipid accumulation in adipocytes and positively regulates adipogenesis. miRNA-144-3p targeting KLF3/carboxy-terminal binding protein 2 (CtBP2) induces C/EBPα activity by releasing corepressors (KLF3 and CtBP2) from its promoter region (80). Ectopic overexpression of miRNA-182 in human adipocytes suppresses the formation of lipid droplets and the expression of adipogenic genes. miRNA-182 belongs to the miRNA-183 cluster and SREBP2 activates miRNA-182 transcription. miRNA-182 targets the 3'-untranslated sequence of C/EBPα directly. miRNA-182 as a novel negative regulator of adipogenesis, may be a potential therapeutic target for obesity in mice with high-fat diet-induced obesity, ob/ob mice, or obese individuals (81). miRNA-200b regulates adipocyte differentiation in 3T3-L1 cells by directly targeting KLF4, and it suppresses the expression of KLF4 in both mRNA level and protein level. All transcription factors are decreased by miRNA-200b overexpression (82). miRNA-144-3p, miRNA-182 and miRNA-200b are thought to be future targets for therapeutic intervention in obesity and metabolic syndrome.
One locus encoding two miRNAs, 378/378*, contained within the intron of PPARγ coactivator 1beta (PGC-1β) is highly induced during adipogenesis in 3T3-L1 cells and ST2 mesenchymal precursor cell lines. miRNA378/378* specifically increases transcriptional activity of C/EBPα and C/EBPβ on adipocyte gene promoters (83). Tumor necrosis factor α (TNF-α) is known to inhibit adipogenesis. TNF-α up-regulates the miRNA-155 through the NF-κB pathway in 3T3-L1 preadipocytes. Thereby, overexpression of miRNA-155 inhibits the expression of C/EBPβ and CREB by directly targeting their 3'UTRs (84). As mentioned above, miRNAs are released into body fluids by adipocytes and can be transported via exosomes to other organs, where they promote ectopic fat deposits by regulating adipogenesis.
Alteration of exosomal miRNA profiles in adipogenesis of human adipose-derived stem cells (hASCs)
Adipogenic stimuli promote cell turnover and synchronized cell division in human multipotent mesenchymal stem cells. This process depends on the stimulation of transcription factors, which are members of the C/EBP family. On the one side adipogenic differentiation is promoted by the major transcriptional inducers of adipogenic gene expression, on the other side inhibitors of adipogenic differentiation, Wingless (Wnt)/β-catenin and the transforming growth factor-β (TGF-β) are repressed. miRNAs are released by adipose tissue, and they are closely associated with the differentiation of adipocytes and are dysregulated in obesity (85). miRNAs regulate this critical signaling pathways related to adipogenesis. They are located within coding DNA sequences such as exons/introns, and across a splice site or in intergenic regions. Just as different miRNA location affects the transcription modality, different miRNAs in the same localization may have opposite effects. Intronic miRNAs use their own promoter, while miRNAs in intergenic regions use the host gene transcriptional machinery and could be transcribed with their host gene, sharing common regulatory patterns (86). Changes in the composition and levels of circulating adipocyte-derived exosomal miRNAs are suggested to be a cause, not a consequence, of obesity in humans with no other health problems. Clinical evidence has shown that AdEXs are present at low levels in the circulation of healthy individuals, in contrast to individuals with obesity (19). Since adipocytes of obese subjects have hypertrophic growth, its cargo content of newly released AdEXs in obese individual reflect the obesity-related metabolic alterations. Number of circulating AdEX is associated with insulin resistance (16).
In a global miRNA expression microarray of 799 miRNAs, 50 miRNAs are significantly differed between either preadipocytes or mature adipocytes, obtained from lean and obese subjects. Seventy miRNAs are highly and significantly up or down-regulated in mature adipocytes as compared to preadipocytes (87). Sixty-five per cent of these differentially expressed miRNAs are located in obesity related chromosomal regions (88). Twenty-three circulating miRNAs are significantly downregulated in diet-induced obesity but upregulated with weight reduction achieved through low-fat diet. These genes are predominantly involved in adipocytokine signaling pathways that are directly associated with obesity and weight reduction (89). Differential expression of 5018 different mRNA probe sets and 15 miRNAs after weight loss indicates that impaired miRNA target gene expression in obese adipose tissue is closely associated with obesity-related inflammation and both recover after weight loss (90). The expression differences have been detected between obese and lean visceral adipose tissue originated exosomes. Thus, lean visceral AdEXs concentration is higher than obese visceral AdEXs (91). Among the miRNAs located in chromosomal regions related to obesity, hsa-miR-15a-5p, hsa-miR-106b-5p, hsa-miR-181a-5p, hsa-let-7 family, hsa-miR-27a-3p, hsa-miR-130b-3p, hsa-miR-152/148a-3p and hsa-miR-26b-5 are particularly important in adipogenesis (92). As mentioned above, the expression levels of miRNAs during the differentiation phase of adipocytes changes after obesity becomes permanent. Furthermore, the miRNA types expressed in adult obesity and prepubertal obesity are different. Thus, Ortega et al. showed that, the expression of 40 miRNAs in preadipocytes and 31 miRNAs in adipocytes are significantly changed in obese individuals. Particularly miRNA-221, miR-222, miR-100 and miR-125b are down-regulated during adipogenesis and proportional with body mass index. In persistent obesity, miRNA-221, miRNA-125b, miRNA-34a, and miRNA-100 are up-regulated, while miRNA-130b, miRNA-210, and miRNA-185 are down-regulated (87). In prepubertal obesity, miRNA-221 and miRNA-28-3p decrease, whereas miRNA-486-5p, miR-486-3p, miR-142-3p, miR-130b, and miR-423-5p increase. Alterations of these miRNAs are significantly correlated with body mass index, as well as other measures of obesity (93). On the other hand, morbidly obese patients show a marked increase in miRNA-140-5p, miRNA-142-3p and miRNA-222. In contrast, the levels of miRNA-532-5p, miRNA-125b, miRNA-130b, miRNA-221, miRNA-15a, miRNA-423-5p, and miRNA-520c-3p decrease (94). Among 30 circulating miRNAs from 155 morbidly obese patients, the most pronounced rise is observed for miRNA-122, miRNA-885-5p, miRNA-148a and miRNA-210, whereas miRNA-150 are markedly decreased. Analysis of relationship between baseline miRNAs levels and clinical variables revealed that the levels of these miRNAs were positively correlated with body mass index and percentage fat mass (95).
Some miRNAs are found in both persistently obese individuals and in adipogenesis. But the functions of these circulating miRNAs have not been fully determined. However, miRNAs that are upregulated in persistent obesity are downregulated during adipogenesis. Moreover, some of the down-regulated miRNAs in obese individuals are up-regulated in mature adipocytes. These findings emphasize that the maturation of undifferentiated adipocytes and adipocyte hypertrophy are different stages of the development of adiposity. In adipose tissue biopsies, the expression of 40 miRNAs in pre-adipocytes and 31 miRNAs in mature adipocytes significantly differ. The difference between the miRNA profiles of human pre-adipocytes and mature adipocytes indicates a close crosstalk between miRNAs and adipogenesis (87).
Visceral adipose tissues of obese and lean individuals differentially express 55 miRNAs, which are related to TGF-β and Wnt/β-catenin signaling pathways, and target 7,789 mRNAs. The expression differences have been detected between obese and lean visceral exosomes for 7 miRNAs. Of these, miRNA-148b and miRNA-4269 are downregulated, while miRNA-23b and miRNA-4429 are upregulated, in the obese subjects. Upregulated miRNAs activate both TGF-β and Wnt/β-catenin signaling pathways in recipient cells (91). Thus, visceral obese adipocytes release the exosomes that contain mediators capable of activating inflammatory and fibrotic signaling pathways. In this regard, visceral adipose tissue sheds exosomes primarily of adipocyte, not macrophage, origin (91). In fact, the overexpression of KLF7 in human preadipocytes suppresses adipogenesis via inhibiting the expression of the adipogenic transcription factors C/EBPα and PPARγ and adipocyte-marker genes. In contrast, inhibition of KLF7 expression like SIRT1, by miR-146b enhances adipogenesis in human visceral adipocytes (96). Kinoshita et al. found that tryptophan hydroxylase-1 (TPH1) is a rate-limiting enzyme for the synthesis of serotonin [5-hydroxytryptamine (5-HT)], and the signaling of its receptors 5-HT2A receptor (5-HT2A R) and 5-HT2C R is involved in adipocyte differentiation. miRNA-448 is encoded in an intronic sequence of the serotonin Htr2c and is in located in the fourth intron of Htr2c. miRNA-448 targets KLF5, which is the binding site of 3'-UTR. Thus, miRNA-448 inhibits adipocyte differentiation by translational repression of KLF5 (97) (Figure 1).
Like miRNA-146b, miRNA-148a, miRNA-26b and miRNA-335 expressions are also dysregulated during the adipocyte differentiation. The levels of miRNA-148a, miRNA-26b, miRNA-30, and miRNA-199a increase during the differentiation of human mesenchymal stem cells (hMSCs)-adipocyte. Among these miRNAs, miRNA-148a exhibits a significant increase in adipogenesis via directly inhibiting the expression of its target gene, Wnt1. There is a positive correlation between adiposity and miRNA-148a expression (98). One of the functional miRNAs during adipogenesis in humans, miR-26b as an intronic one, localizes in the intron of carboxy terminal domain, RNA polymerase II, polypeptide A, small phosphatase 1 (CTDSP1) of human adipose-derived mesenchymal stem cell (hADMSC) and is gradually upregulated during adipocyte differentiation. Bioinformatics analyses showed that the target of this miRNA is the Wnt/β-catenin pathway component T cell factor 4. Adipogenesis is effectively suppressed as Wnt/β-catenin is activated when miRNA-26b is inhibited (99).
miRNA-27a is an intergenic miRNA, whereas miRNA-27b is an intronic miRNA. During the adipogenic differentiation of hASCs, miRNA-27a which is extracted from the human adipose tissues, is more abundantly expressed, miR-27b expression is down-regulated. miRNA-27b increases adipogenesis of hASCs, whereas miRNA-27a inhibits adipogenesis by decreasing the expression of two key transcription factors, PPARγ and C/EBPα (47,100). Another miRNA that changes during adipocyte differentiation is miRNA-335, which affects both fatty acid metabolism and lipogenesis. If human mature adipocytes are exposed to adipokines, increases in miRNA-335 expression leads to adipocyte differentiation (101). Pri-miRNA-21 is located within the intronic region and is independently transcribed by its own promoter regions. Processed miRNA-21 expression is transiently increased after induction of adipogenic differentiation of hASCs, peaked at initial phase. The level of miRNA-21 expression is 10-fold higher than that of miRNA-143 expression. Increased expression of miRNA-21 decreases both protein and mRNA levels of miRNA-21 targets genes. Thus, TGF-beta-induced inhibition of adipogenic differentiation is significantly decreased in miRNA-21 expressing cells (102).
Overexpression of miRNA-17-5p in response to high fat diet activates adipogenesis via increasing adipogenic transcription factors and repressing the Wnt signaling pathway. Thus, miRNA-17-5p is among the central switches of adipogenic differentiation (103). However, expression of miRNA-17-5p is significantly different between obese and nonobese omental fat (104). Highly induced miRNA378/378* during adipogenesis increases transcriptional activity of C/EBPα and C/EBPβ on adipocyte gene promoters in cell lines and enhances adipogenesis (83). In the same way, brown adipocyte-derived exosomal miRNA-378 is a positive regulator of adipogenesis in constitutive brown adipose tissue (cBAT) of human. Its overexpression leads to cBAT expansion, resulting in increased energy expenditure, and protects against genetically determined diet-induced obesity (105). Levels of this miRNA is also significantly over-expressed during the normal adipogenesis in humans and directly correlated with adipogenic transcription factors (87).
Like miRNA-27b, miRNA-138 is significantly down-regulated during adipogenic differentiation. Human miRNA, hsa-miRNA-138 inhibits the adipogenic differentiation of hAD-MSCs. While overexpression of miRNA-138 in hAD-MSCs effectively reduces lipid droplets accumulation, miRNA inhibits expression of both key adipogenic transcription factors C/EBPα and PPARγ (106). Adipogenic differentiation of human mesenchymal stromal cells is impaired by miRNA-369-5p whereas it is highly increased by miRNA-371. This is accompanied by respective gene expression changes of adiponectin and FABP4 (107). Obese women have significantly lower miRNA-130 and higher PPARγ mRNA levels than that from nonobese women. miRNA-130 strongly affects adipocyte differentiation and reduces adipogenesis by repressing PPARγ biosynthesis. Interestingly, repression of miRNA-130 on PPARγ expression happen via targeting both the PPARγ mRNA coding and 3'UTR (108).
Conclusions
In obesity, adipogenesis is controlled by coordinated actions of transcription factors and epigenomic regulators. In the last decade, an increasing number of studies have identified novel transcriptional factors and epigenomic regulators of adipogenesis. Of these “key epigenomic regulators”, miRNAs play opposing roles in the regulation of adipogenesis. Thus, miRNAs can accelerate or inhibit adipocyte differentiation by affecting transcription factors and regulating signaling pathways related to adipogenesis. Therefore, differential expression of miRNAs during adipogenesis and permanent obesity allows their use as biomarkers or therapeutic targets. It is expected that miRNA-based therapeutics targeting obesity will be a tool for the methods to be developed in the treatment of obesity. However, the ability of a miRNA to interact with many targets, combined with the possibility for several miRNAs sharing the same target, constitute diverse regulatory mechanisms that tremendously increase the complexity of biological networks (Figure 1). Thus, the broad-spectrum and redundancy of miRNA-target interactions constitute a major challenge in the development of miRNA-targeted therapeutics.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Chenyu Zhang) for the series “Extracellular RNAs and Human Health” published in ExRNA. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-4/coif). The series “Extracellular RNAs and Human Health” was commissioned by the editorial office without any funding or sponsorship. AE serves as an unpaid editorial board member of ExRNA from November 2021 to October 2023. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leung MY, Pollack LM, Colditz GA, et al. Life years lost and lifetime health care expenditures associated with diabetes in the U.S., National Health Interview Survey, 1997-2000. Diabetes Care 2015;38:460-8. [Crossref] [PubMed]
- Engin A. The Definition and Prevalence of Obesity and Metabolic Syndrome. Adv Exp Med Biol 2017;960:1-17. [Crossref] [PubMed]
- Kramer CK, Zinman B, Retnakaran R. Are metabolically healthy overweight and obesity benign conditions?: A systematic review and meta-analysis. Ann Intern Med 2013;159:758-69. [Crossref] [PubMed]
- Blüher M. Are there still healthy obese patients? Curr Opin Endocrinol Diabetes Obes 2012;19:341-6. [Crossref] [PubMed]
- Porro S, Genchi VA, Cignarelli A, et al. Dysmetabolic adipose tissue in obesity: morphological and functional characteristics of adipose stem cells and mature adipocytes in healthy and unhealthy obese subjects. J Endocrinol Invest 2021;44:921-41. [Crossref] [PubMed]
- Engin AB. What Is Lipotoxicity? Adv Exp Med Biol 2017;960:197-220. [Crossref] [PubMed]
- Rodriguez AM, Elabd C, Delteil F, et al. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem Biophys Res Commun 2004;315:255-63. [Crossref] [PubMed]
- Zuk PA, Zhu M, Ashjian P, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 2002;13:4279-95. [Crossref] [PubMed]
- Spalding KL, Arner E, Westermark PO, et al. Dynamics of fat cell turnover in humans. Nature 2008;453:783-7. [Crossref] [PubMed]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006;7:885-96. [Crossref] [PubMed]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 2011;12:722-34. [Crossref] [PubMed]
- Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol 2000;16:145-71. [Crossref] [PubMed]
- Engin AB. MicroRNA and Adipogenesis. Adv Exp Med Biol 2017;960:489-509. [Crossref] [PubMed]
- Baulcombe D. RNA silencing. Trends Biochem Sci 2005;30:290-3. [Crossref] [PubMed]
- Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature 2005;434:338-45. [Crossref] [PubMed]
- Eguchi A, Lazic M, Armando AM, et al. Circulating adipocyte-derived extracellular vesicles are novel markers of metabolic stress. J Mol Med (Berl) 2016;94:1241-53. [Crossref] [PubMed]
- Engin A, Engin AB. Why should the molecular characterization of inflammasome-induced exosomal cargo be done? ExRNA 2021;3:6. [Crossref]
- Rome S, Blandin A, Le Lay S. Adipocyte-Derived Extracellular Vesicles: State of the Art. Int J Mol Sci 2021;22:1788. [Crossref] [PubMed]
- Zhou Y, Tan C. miRNAs in Adipocyte-Derived Extracellular Vesicles: Multiple Roles in Development of Obesity-Associated Disease. Front Mol Biosci 2020;7:171. [Crossref] [PubMed]
- Li X, Ballantyne LL, Yu Y, et al. Perivascular adipose tissue-derived extracellular vesicle miR-221-3p mediates vascular remodeling. FASEB J 2019;33:12704-22. [Crossref] [PubMed]
- Dang SY, Leng Y, Wang ZX, et al. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int J Biol Sci 2019;15:351-68. [Crossref] [PubMed]
- Kim MS, Muallem S, Kim SH, et al. Exosomal release through TRPML1-mediated lysosomal exocytosis is required for adipogenesis. Biochem Biophys Res Commun 2019;510:409-15. [Crossref] [PubMed]
- Connolly KD, Guschina IA, Yeung V, et al. Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J Extracell Vesicles 2015;4:29159. [Crossref] [PubMed]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993;73:725-34. [Crossref] [PubMed]
- Crandall DL, Hausman GJ, Kral JG. A review of the microcirculation of adipose tissue: anatomic, metabolic, and angiogenic perspectives. Microcirculation 1997;4:211-32. [Crossref] [PubMed]
- Gealekman O, Guseva N, Hartigan C, et al. Depot-specific differences and insufficient subcutaneous adipose tissue angiogenesis in human obesity. Circulation 2011;123:186-94. [Crossref] [PubMed]
- Pasarica M, Sereda OR, Redman LM, et al. Reduced adipose tissue oxygenation in human obesity: evidence for rarefaction, macrophage chemotaxis, and inflammation without an angiogenic response. Diabetes 2009;58:718-25. [Crossref] [PubMed]
- Sano S, Izumi Y, Yamaguchi T, et al. Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochem Biophys Res Commun 2014;445:327-33. [Crossref] [PubMed]
- Engin A. Adipose Tissue Hypoxia in Obesity and Its Impact on Preadipocytes and Macrophages: Hypoxia Hypothesis. Adv Exp Med Biol 2017;960:305-26. [Crossref] [PubMed]
- Wahid F, Shehzad A, Khan T, et al. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 2010;1803:1231-43. [Crossref] [PubMed]
- O'Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 2013;38:39-54. [Crossref] [PubMed]
- Wei Y, Corbalán-Campos J, Gurung R, et al. Dicer in Macrophages Prevents Atherosclerosis by Promoting Mitochondrial Oxidative Metabolism. Circulation 2018;138:2007-20. [Crossref] [PubMed]
- Mang GM, Pradervand S, Du NH, et al. A neuron-specific deletion of the microRNA-processing enzyme DICER induces severe but transient obesity in mice. PLoS One 2015;10:e0116760. [Crossref] [PubMed]
- Schneeberger M, Altirriba J, García A, et al. Deletion of miRNA processing enzyme Dicer in POMC-expressing cells leads to pituitary dysfunction, neurodegeneration and development of obesity. Mol Metab 2012;2:74-85. [Crossref] [PubMed]
- Vinnikov IA, Hajdukiewicz K, Reymann J, et al. Hypothalamic miR-103 protects from hyperphagic obesity in mice. J Neurosci 2014;34:10659-74. [Crossref] [PubMed]
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009;58:1050-7. [Crossref] [PubMed]
- Wen Z, Li J, Fu Y, et al. Hypertrophic Adipocyte-Derived Exosomal miR-802-5p Contributes to Insulin Resistance in Cardiac Myocytes Through Targeting HSP60. Obesity (Silver Spring) 2020;28:1932-40. [Crossref] [PubMed]
- Müller G, Schneider M, Biemer-Daub G, et al. Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell Signal 2011;23:1207-23. [Crossref] [PubMed]
- McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med 2011;11:304-16. [Crossref] [PubMed]
- Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert Opin Ther Targets 2009;13:1227-38. [Crossref] [PubMed]
- Ahn J, Lee H, Jung CH, et al. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol Med 2013;5:1602-12. [Crossref] [PubMed]
- Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006;127:1109-22. [Crossref] [PubMed]
- Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004;429:771-6. [Crossref] [PubMed]
- Zhu YL, Chen T, Xiong JL, et al. miR-146b Inhibits Glucose Consumption by Targeting IRS1 Gene in Porcine Primary Adipocytes. Int J Mol Sci 2018;19:783. [Crossref] [PubMed]
- Wu D, Xi QY, Cheng X, et al. miR-146a-5p inhibits TNF-α-induced adipogenesis via targeting insulin receptor in primary porcine adipocytes. J Lipid Res 2016;57:1360-72. [Crossref] [PubMed]
- Oishi Y, Manabe I, Tobe K, et al. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 2005;1:27-39. [Crossref] [PubMed]
- Kim SY, Kim AY, Lee HW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 2010;392:323-8. [Crossref] [PubMed]
- Wildwater M, Sander N, de Vreede G, et al. Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 2011;138:4375-85. [Crossref] [PubMed]
- Qin L, Chen Y, Niu Y, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics 2010;11:320. [Crossref] [PubMed]
- Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004;279:52361-5. [Crossref] [PubMed]
- Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 2006;12:1626-32. [Crossref] [PubMed]
- An X, Ma K, Zhang Z, et al. miR-17, miR-21, and miR-143 Enhance Adipogenic Differentiation from Porcine Bone Marrow-Derived Mesenchymal Stem Cells. DNA Cell Biol 2016;35:410-6. [Crossref] [PubMed]
- Wang L, Zhang S, Cheng G, et al. MiR-145 reduces the activity of PI3K/Akt and MAPK signaling pathways and inhibits adipogenesis in bovine preadipocytes. Genomics 2020;112:2688-94. [Crossref] [PubMed]
- Chung JY, Hong J, Kim HJ, et al. White adipocyte-targeted dual gene silencing of FABP4/5 for anti-obesity, anti-inflammation and reversal of insulin resistance: Efficacy and comparison of administration routes. Biomaterials 2021;279:121209. [Crossref] [PubMed]
- Ling HY, Wen GB, Feng SD, et al. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin Exp Pharmacol Physiol 2011;38:239-46. [Crossref] [PubMed]
- Bost F, Aouadi M, Caron L, et al. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005;87:51-6. [Crossref] [PubMed]
- Accili D, Taylor SI. Targeted inactivation of the insulin receptor gene in mouse 3T3-L1 fibroblasts via homologous recombination. Proc Natl Acad Sci U S A 1991;88:4708-12. [Crossref] [PubMed]
- Miki H, Yamauchi T, Suzuki R, et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol 2001;21:2521-32. [Crossref] [PubMed]
- Tseng YH, Kriauciunas KM, Kokkotou E, et al. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol 2004;24:1918-29. [Crossref] [PubMed]
- Lin W, Tang Y, Zhao Y, et al. MiR-144-3p Targets FoxO1 to Reduce Its Regulation of Adiponectin and Promote Adipogenesis. Front Genet 2020;11:603144. [Crossref] [PubMed]
- Li Y, Ma Z, Jiang S, et al. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog Lipid Res 2017;66:42-9. [Crossref] [PubMed]
- Dong P, Mai Y, Zhang Z, et al. MiR-15a/b promote adipogenesis in porcine pre-adipocyte via repressing FoxO1. Acta Biochim Biophys Sin (Shanghai) 2014;46:565-71. [Crossref] [PubMed]
- Zhou Z, Tao Y, Zhao H, et al. Adipose Extracellular Vesicles: Messengers From and to Macrophages in Regulating Immunometabolic Homeostasis or Disorders. Front Immunol 2021;12:666344. [Crossref] [PubMed]
- Ogawa R, Tanaka C, Sato M, et al. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem Biophys Res Commun 2010;398:723-9. [Crossref] [PubMed]
- Engin AB. Adipocyte-Macrophage Cross-Talk in Obesity. Adv Exp Med Biol 2017;960:327-43. [Crossref] [PubMed]
- Zhang Y, Mei H, Chang X, et al. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J Mol Cell Biol 2016;8:505-17. [Crossref] [PubMed]
- Essandoh K, Li Y, Huo J, et al. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016;46:122-31. [Crossref] [PubMed]
- Dang CP, Leelahavanichkul A. Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PLoS One 2020;15:e0236038. [Crossref] [PubMed]
- Pan Y, Hui X, Hoo RLC, et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest 2019;129:834-49. [Crossref] [PubMed]
- Cho YK, Son Y, Kim SN, et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol Metab 2019;29:86-98. [Crossref] [PubMed]
- Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev 1996;10:1096-107. [Crossref] [PubMed]
- Fernández-Hernando C, Ramírez CM, Goedeke L, et al. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 2013;33:178-85. [Crossref] [PubMed]
- Gerin I, Clerbaux LA, Haumont O, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 2010;285:33652-61. [Crossref] [PubMed]
- Price NL, Holtrup B, Kwei SL, et al. SREBP-1c/MicroRNA 33b Genomic Loci Control Adipocyte Differentiation. Mol Cell Biol 2016;36:1180-93. [Crossref] [PubMed]
- Cirera-Salinas D, Pauta M, Allen RM, et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 2012;11:922-33. [Crossref] [PubMed]
- Sun T, Fu M, Bookout AL, et al. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol 2009;23:925-31. [Crossref] [PubMed]
- Rice SJ, Lai SC, Wood LW, et al. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J Biol Chem 2013;288:16348-60. [Crossref] [PubMed]
- Rosen ED, Hsu CH, Wang X, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002;16:22-6. [Crossref] [PubMed]
- Zhou J, Yang J, Wang X, et al. A Novel Regulatory Circuit "C/EBPα/miR-20a-5p/TOB2" Regulates Adipogenesis and Lipogenesis. Front Endocrinol (Lausanne) 2020;10:894. [Crossref] [PubMed]
- Shen L, Li Q, Wang J, et al. miR-144-3p Promotes Adipogenesis Through Releasing C/EBPα From Klf3 and CtBP2. Front Genet 2018;9:677. [Crossref] [PubMed]
- Dong M, Ye Y, Chen Z, et al. MicroRNA 182 is a Novel Negative Regulator of Adipogenesis by Targeting CCAAT/Enhancer-Binding Protein α. Obesity (Silver Spring) 2020;28:1467-76. [Crossref] [PubMed]
- Shen L, Gan M, Li Q, et al. MicroRNA-200b regulates preadipocyte proliferation and differentiation by targeting KLF4. Biomed Pharmacother 2018;103:1538-44. [Crossref] [PubMed]
- Gerin I, Bommer GT, McCoin CS, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab 2010;299:E198-206. [Crossref] [PubMed]
- Liu S, Yang Y, Wu J. TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem Biophys Res Commun 2011;414:618-24. [Crossref] [PubMed]
- Ji C, Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol 2019;15:731-43. [Crossref] [PubMed]
- Monteys AM, Spengler RM, Wan J, et al. Structure and activity of putative intronic miRNA promoters. RNA 2010;16:495-505. [Crossref] [PubMed]
- Ortega FJ, Moreno-Navarrete JM, Pardo G, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One 2010;5:e9022. [Crossref] [PubMed]
- Rankinen T, Zuberi A, Chagnon YC, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529-644. [Crossref] [PubMed]
- Hsieh CH, Rau CS, Wu SC, et al. Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC Genomics 2015;16:699. [Crossref] [PubMed]
- Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Surgery-Induced Weight Loss Is Associated With the Downregulation of Genes Targeted by MicroRNAs in Adipose Tissue. J Clin Endocrinol Metab 2015;100:E1467-76. [Crossref] [PubMed]
- Ferrante SC, Nadler EP, Pillai DK, et al. Adipocyte-derived exosomal miRNAs: a novel mechanism for obesity-related disease. Pediatr Res 2015;77:447-54. [Crossref] [PubMed]
- Shi C, Huang F, Gu X, et al. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 2016;7:40830-45. [Crossref] [PubMed]
- Prats-Puig A, Ortega FJ, Mercader JM, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab 2013;98:E1655-60. [Crossref] [PubMed]
- Ortega FJ, Mercader JM, Catalán V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem 2013;59:781-92. [Crossref] [PubMed]
- Sangiao-Alvarellos S, Theofilatos K, Barwari T, et al. Metabolic recovery after weight loss surgery is reflected in serum microRNAs. BMJ Open Diabetes Res Care 2020;8:e001441. [Crossref] [PubMed]
- Chen L, Dai YM, Ji CB, et al. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol Cell Endocrinol 2014;393:65-74. [Crossref] [PubMed]
- Kinoshita M, Ono K, Horie T, et al. Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol Endocrinol 2010;24:1978-87. [Crossref] [PubMed]
- Shi C, Zhang M, Tong M, et al. miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci Rep 2015;5:9930. [Crossref] [PubMed]
- Song G, Xu G, Ji C, et al. The role of microRNA-26b in human adipocyte differentiation and proliferation. Gene 2014;533:481-7. [Crossref] [PubMed]
- Karbiener M, Fischer C, Nowitsch S, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun 2009;390:247-51. [Crossref] [PubMed]
- Zhu L, Chen L, Shi CM, et al. MiR-335, an adipogenesis-related microRNA, is involved in adipose tissue inflammation. Cell Biochem Biophys 2014;68:283-90. [Crossref] [PubMed]
- Kim YJ, Hwang SJ, Bae YC, et al. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009;27:3093-102. [Crossref] [PubMed]
- Tian L, Song Z, Shao W, et al. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis 2017;8:e2559. [Crossref] [PubMed]
- Heneghan HM, Miller N, McAnena OJ, et al. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab 2011;96:E846-50. [Crossref] [PubMed]
- Pan D, Mao C, Quattrochi B, et al. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat Commun 2014;5:4725. [Crossref] [PubMed]
- Yang Z, Bian C, Zhou H, et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev 2011;20:259-67. [Crossref] [PubMed]
- Bork S, Horn P, Castoldi M, et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol 2011;226:2226-34. [Crossref] [PubMed]
- Lee EK, Lee MJ, Abdelmohsen K, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011;31:626-38. [Crossref] [PubMed]
Cite this article as: Engin AB, Engin A. Adipogenesis-related microRNAs in obesity. ExRNA 2022;4:16.


