
DodecaRNAs (doRNAs) are abundant in cow’s milk and differentially enriched in milk ultracentrifugation fractions
Introduction
Extracellular RNAs (exRNAs) are found in a diversity of biological fluids, but they are especially enriched in milk (1,2). Within this fluid, exRNAs are contained in small membranous extracellular vesicles (EVs), such as exosomes, that protect their labile cargo of exRNAs (3,4) from the degradative conditions that prevail during lactation (e.g., RNase found in milk, IgA hydrolases) (5,6) and digestion [(7) and reviewed in (8,9)].
For the last decade, most studies investigating milk exRNAs in different species focused on microRNAs (4,9-12), small non-coding RNAs (ncRNAs) implicated in a vast array of—if not all—physiological functions and pathologies, such as cancer and inflammatory diseases [reviewed in (13)]. However, our previous discovery of a plethora of milk EV (mEV) subsets (6,14), progress of small RNA sequencing technologies and democratization of their use initiated new lines of research unveiling the relative abundance and diversity of other non-coding exRNAs, such as ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), tRNA fragments (tRFs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs) (10,15,16) and coding exRNA (i.e., mRNAs) (17,18).
Recently, the discovery of very short [12–13 nucleotides; (nt)] functional dodecaRNAs (doRNAs) (19) challenged the belief that ncRNAs shorter than 16 nt were merely non-functional degradative byproducts that pollute sequencing results. Most probably derived from the 5.8S rRNA, doRNAs were found to be extremely abundant in the cells, organs and even EVs (i.e., platelet-derived EVs) that were analyzed. Found so far in humans, mice and flies (19), doRNAs displayed a species-specific enrichment and ratio of the two most abundant isomers of doRNAs: the 12-nt doRNA and its 13-nt c-doRNA [a doRNA derivative harboring an additional cytosine (C) at its 5’ end] variant (19). Moreover, these very short ncRNAs were notably found to be more abundant than microRNAs in these samples. Interestingly, they were mainly cytoplasmic, interacted with heterogeneous nuclear ribonucleoproteins (hnRNP) A0, A1 and A2B1, and were found to regulate the expression of Annexin II receptor (AXIIR) (19). They were also differentially expressed in prostate cancer cells/tissues (vs healthy controls) and impacted cancer cell migration (19). Along with our previous report on semi-microRNAs (smiRNAs) (20), these results put in question the practice of systematically discarding sequencing data of RNAs shorter than the arbitrary threshold of 16 nt, oftentimes even before library construction (21).
While we previously reported the existence of different RNA biotypes, including canonical rRNA fragments (rRFs) longer than 16 nt in commercial dairy milk (16), whether shorter RNA species derived from rRNA, such as doRNAs, exist in milk remains unknown. Because (I) milk is highly enriched in exRNAs of various types, (II) milk exRNAs resist human digestion (7) and (III) milk exRNAs might exert biological functions (12,22) and impact consumers’ health upon ingestion [reviewed in (8,11)], we hypothesized that commercially available dairy cow’s milk might contain doRNAs or similar, very short exRNAs, in greater amount than microRNAs.
To fill this gap in knowledge and after an extensive characterization of mEVs (6,7,14,16,23), we used small RNA-sequencing (sRNA-seq) to investigate the existence and abundance of 8- to 15-nt sRNAs, including doRNAs, in commercial dairy cow’s milk and validated these results using a new high-specificity reverse transcription quantitative polymerase chain reaction (RT-qPCR) method designed and validated for RNAs shorter than microRNAs (24). We present the following article in accordance with the MDAR reporting checklist (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-6/rc).
Methods
Cow milk samples
Commercial skimmed filtered dairy milk (PurFiltre, Lactantia, Toronto, Canada) was purchased in a local store in Québec City, Canada. Three packs of milk with different expiration dates were either processed for triplicate analyses (sequencing of unfractionated milk and qPCR validations) or were mixed into one milk solution used for sequencing analyses of milk fractions.
Differential ultracentrifugation
Milk was fractioned following our previously reported protocol (14). Briefly, to solubilize casein and prevent contamination of the mEVs-enriched fractions with this protein, 100 mL of dairy milk was mixed with 1 volume of 2% sodium citrate (in water, Sigma) filtered through 0.22-µm pore microfilters (Corning, Corning, NY, USA) and kept on a rocking table for 20 min at 4 °C until milk clarified. The citrated milk samples were then subjected to successive ultracentrifugation at 12,000 (12K), 35,000 (35K), 70,000 (70K) and 100,000 (100K) g for 1 h each at 4 °C in a Sorvall WX TL-100 ultracentrifuge, equipped with either a SureSpin 630 or a T-1250 Rotor (Sorvall, through Thermo Fisher Scientific, Waltham, MA, USA). After each step, pellets were carefully suspended in 1 mL of 0.22-µm filtered sterile phosphate buffer saline (PBS), pH 7.4 and kept resuspending overnight at 4 °C before RNA isolation.
RNA isolation
For sequencing analyses, total RNA from milk or milk fractions was isolated using TRIzol LS (Thermo Fisher Scientific) following the manufacturer’s recommendations prior to resuspension in diethylpyrocarbonate (DEPC)-treated, nuclease-free water (Invitrogen, Carlsbad, CA, USA). Total RNA was purified further using RNeasy mini-kit and subjected to on-column treatment with DNase I, according to the manufacturer’s protocol (Qiagen, Hilden, Germany). Total RNAs was kept at −80 °C for a few days, after which it was shipped on dry ice to the ArrayStar sequencing platform (Rockville, MD, Canada).
For qPCR analyses, total RNA was isolated from 250 µL of whole milk or resuspended mEVs after mixing with 750 µL TRIzol LS (Thermo Fisher Scientific) spiked with UniSp2 exogenous synthetic RNA oligonucleotide (Qiagen, MD, USA, Cat No. 339306, product number: YP00203950), used as an internal control, following the manufacturer’s recommendations. Isolated RNA was further treated with DNase I (M0303S, New England Biolabs, MA, USA), as per manufacturer’s protocol, to remove contaminating DNA. RNA was stored at −80 °C for a few days before reverse transcription.
Complementary DNA (cDNA) library preparation
Library preparations were performed following modified standard operating procedures (SOPs) at ArrayStar platform to allow for a larger RNA size window (8–35 nt). The purity, quality, and concentration of total RNA samples were determined with NanoDrop ND-1000 (Thermo Fisher Scientific) and 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The miRNA sequencing library was prepared from total RNA through: (I) 3'-adapter ligation; (II) 5'-adapter ligation; (III) cDNA synthesis; (IV) PCR amplification, and (V) size selection of approximately 120 to 155 bp of PCR-amplified fragments (corresponding to approximately 8 to 35 nt of small RNA). The complete libraries were analyzed and quantitated by Agilent 2100 Bioanalyzer.
sRNA-seq
The cDNA library samples were then diluted to a final concentration of 8 pM and denatured as single-stranded DNA. Cluster generation was performed on the Illumina cBot using TruSeq Rapid SR cluster kit (#GD-402–4001, Illumina, San Diego, CA, USA) and sequenced for 51 cycles on Illumina HiSeq 2000, using TruSeq Rapid SBS Kits (#FC-402–4002, Illumina), following the manufacturer’s instructions.
Bioinformatics analysis of sequencing data
Adapter sequences were then trimmed from the reads that passed the quality control filter (clean reads) leaving clean sRNA trimmed reads. All analyses displayed here were provided through the ArrayStar standard analysis pipeline and refined using R (R Foundation for Statistical Computing, Vienna, Austria). Only the reads that were identical, both in length and sequence, were considered as a unique read. Small RNA biotypes were determined by mapping trimmed reads against bovine noncoding RNA database [Bos_taurus.UMD3.1.ncrna, http://bovinegenome.org/; Elsik et al., 2016 (25)] using Blastn tool (National Library of Medicine, National Center for Biotechnology. Information, https://blast.ncbi.nlm.nih.gov/Blast.cgi). For miRNA, trimmed reads were aligned to miRBase pre-microRNA database (miRBase release 22.1, http://www.mirbase.org/). Sequences known to be contaminant confounders from RNA isolation procedures were discarded [Heintz-Buschart et al., 2018 (26)]. sRNA read counts were expressed as reads per million (RPM) sRNAs within the specified size windows.
Adapter-ligated RT-qPCR
The splint bridging sequence and adapter used to quantify sRNAs were annealed together, as described in our previously published protocol (24). The annealed adapter/splint was added to 200 ng of total RNAs (10 µL) and 18 µL of ligation master mix containing 10 units T4 RNA ligase (New England Biolabs, M0437M, Whitby, Ontario, Canada), 10% dimethyl sulfoxide (DMSO), 25% polyethylene glycol (PEG) 8000 (New England Biolabs, Whitby, Ontario, Canada), 1 mM adenosine triphosphate (ATP) (New England Biolabs, M0437M, Whitby, Ontario, Canada) and 20 units SUPERase•In RNase inhibitor (Thermo Fisher Scientific, AM2694, Waltham, MA, USA) in T4 RNA ligase reaction buffer [50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), pH 7.5]. The ligation (in 31 µL) was allowed to proceed for 1 h at room temperature (RT), after which 17 µL of stop ligation buffer [1 M Tris-HCl, 0.1 M ethylene diamine tetraacetic acid (EDTA), pH 8.0] was added. This step lengthened doRNA and c-doRNA to 22 and 23 nt RNAs, respectively. Two µL of ligated total RNA, along with UniSp6 RT spike-in, were used for RT using the miRCURY locked nucleic acid (LNA)-modified microRNA PCR Assay (QIAGEN Inc., Toronto, ON, Canada) and the oligo-d(T) primer with 5’ universal tag included in the miRCURY LNA RT Kit (QIAGEN Inc., Toronto, ON, Canada; Cat. No. #339340). After cDNA 1/10 dilution, qPCR was performed using miRCURY LNA SYBR® Green PCR Kits (QIAGEN Inc., Toronto, ON, Canada) in 0.1 mL MicroAmpTM Fast Optical 96-Well Reaction Plate (Applied BiosystemTM, Cat. No. 4346907) StepOneTM Real-Time PCR System (Cat. No. 4376357) and specific Custom LNA Oligonucleotides for the doRNA (No. 339317, ad3-d-621278, Cat. No. YCP0054421, QIAGEN Inc., Toronto, ON, Canada) and c-doRNA (No. 339317, ad3-C-d-621381_1, Cat. No. YCP0054420, QIAGEN Inc., Toronto, ON, Canada) or the LNA PCR primer assays (QIAGEN Inc., Toronto, ON, Canada) for miR-30a-5p (No. 339306, miR-30a-5p, Cat. No. YP02104140), miR-148a-3p (miR-148a-3p, Cat. No. YP00205867) and Let-7b-5p (Let-7b-5p, Cat. No. YP00204750). We used the following thermal PCR cycle program: denaturation step at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/elongation at 57 °C for 1 min.
Standard curve for absolute doRNAs and miRNAs quantification
DoRNA, c-doRNA, miR-30a-5p, miR-148a-3p and Let-7b-5p copy numbers were determined using a standard curve established using the corresponding synthetic RNA oligonucleotides (IDT, Coralville, IA, USA) serially diluted 1/10th to obtain between 5.7×109 and 5.7×103 copies, covering a range of concentration of 6 logs (Figure S1). Diluted synthetic RNA was then subjected to adapter-ligated RT-qPCR. For each standard curve, the cycle quantitation (Cq) values with the corresponding copy numbers were plotted, and the linear curve equation and correlation coefficient (R2) were calculated (Figure S2). doRNA and miRNA quantifications were normalized using internal spike-in controls to ensure comparable isolation efficiency (isolation and splint-RT-qPCR quality controls available in Figure S3).
Statistical analysis
All statistical analyses were performed using Prism 9.2.0 (GraphPad Software Inc., La Jolla, CA, USA). Unfractionated milk and validation experiments were performed in biological triplicates (n=3). Statistical significance was determined by one-way ANOVA with Holm-Sidak’s post-hoc correction for multiple comparisons or Student’s t-test after validation of statistical assumptions and prerequisite for each test and with type error α set to 0.05 (i.e., 5%, P value below 0.05 considered significant).
Illustrations
Figures were generated using R (R Foundation for Statistical Computing), Inkscape software (http://inkscape.org/), InteractiVenn (27) and Prism 7 (GraphPad Software Inc.).
Results
doRNAs are highly abundant in commercial cow’s milk
There was a higher abundance of sRNAs in the 8–15 nt compared to the 15–30 nt size window of our unfractionated commercial cow’s milk sRNA sequencing data, although this 1.4-fold difference did not reach statistical significance (Figure 1A).
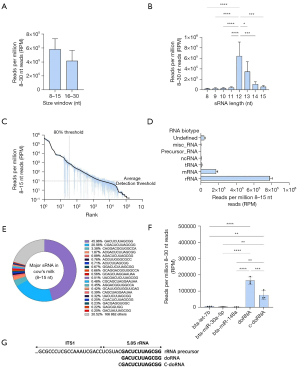
Analysis of size distribution of sRNAs in the 8–15 window revealed a marked, and significant, 6- to 33-fold enrichment of 12-nt sRNAs and 3- to 17-fold enrichment of 13-nt sRNA, in comparison to the other fractions (P<0.05 to P<0.0001, Figure 1B). The sRNA detection threshold in this experiment averaged 10.06 RPM (Figure 1C), with 34±21 sRNAs representing 80% of the entire 49,612 detected sequences (Figure 1C). Most of the sRNAs in these samples mapped to rRNAs (77.9%) and mRNAs (17.1%, Figure 1D). Among the 20 most expressed sRNAs, two sequences (GACUCUUAGCGG and CGACUCUUAGCGG, 12 and 13 nt in length, respectively) represented 66.52% of all 8–15 nt sequences (Figure 1E). These two rRFs were designated as doRNAs, in reference to the number of core nucleotides (12 nt) they contain.
DoRNAs and c-doRNAs were respectively 25 to 49 and 11 to 30 times more enriched in these milk sequencing data than bta-let-7b, bta-miR-30a-5p and bta-miR-148a; three of the most enriched milk microRNAs (28) from the same sequencing datasets (P<0.01 to P<0.0001, Figure 1F). These two sequences are the doRNA and c-doRNA we previously reported in other species (19) and derive most likely from the 5.8S rRNA (Figure 1F,1G).
These results support the existence and high enrichment of the very short doRNAs in cow’s milk, less diverse but more abundant than the most abundant milk microRNAs.
Milk ultracentrifugation fractions have specific 8–15 nt sRNA enrichment profiles
In our previous reports, we fully characterized the mEVs contained in the 12K, 35K, 70K and 100K g fractions obtained upon sequential ultracentrifugation of commercial cow’s milk (6,16,23). We replicated the above-described analysis on these isolated fractions, as described in the Methods sections (Figure 2).
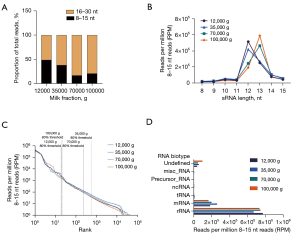
In all fractions, and contrary to unfractionated milk, most sequences (51.3% to 83%) were in the 15–30 nt size window (Figure 2A). There was a fraction-specific ratio between 8–15 and 15–30 nt sRNAs, with an increasing proportion of 16–30 nt sRNAs in the later fractions (70K and 100K g, Figure 2A). Similarly, 12K and 35K g milk fractions had higher proportion of 12-nt sRNAs, while subsequent fractions were more enriched in 13-nt sRNAs (Figure 2B).
Detection thresholds and total sRNA count in these samples were comparable (Figure 2C). In 12K and 100K g fractions, 19 and 20 sequences represented 80% of 8–15 nt reads, respectively (Figure 2C). In 35K and 70K g fractions, this proportion accounted for 562 and 230 sequences, respectively, suggesting a higher diversity of sequences in these two intermediate fractions (Figure 2C). As for unfractionated milk, biotype analysis revealed that most of the 8–15 nt sRNAs, as in all fractions, mapped to rRNAs (73–77%) and mRNAs (16–25%) (Figure 2D).
Further comparison of the four milk fractions revealed that each contained between 21,008 to 37,204 specific sequences and shared over 8,326 common reads (Figure 3A). Enrichment distribution of the 200 most enriched 8–15 nt sRNAs in milk (~80% of all sequences) revealed that those were most enriched in the 12K and 35K g fractions (Figure 3B). Clustering by these sRNAs confirmed 12K and 35K g fractions were more closely related to each other (Figure 3B). Similar observation was drawn for 70K and 100K g fractions (Figure 3B).
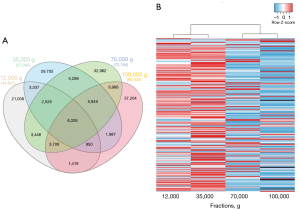
These results suggest 8–15 nt sRNAs, a high proportion of which is around 12–13 nt in size, distribute differentially between milk ultracentrifugation fractions, distinguishing and clustering these in two subsets (12K–35K and 70K–100K g groups).
The majority of 8–15 nt sRNAs in milk fractions are doRNAs that are more abundant than most abundant milk microRNAs
Like unfractionated milk, doRNA (GACUCUUAGCG, 12 nt) and c-doRNA (CGACUCUUAGCGCC, 13 nt) accounted for 56% to 67% of all 8–15 sRNAs (Figure 4A-4D).
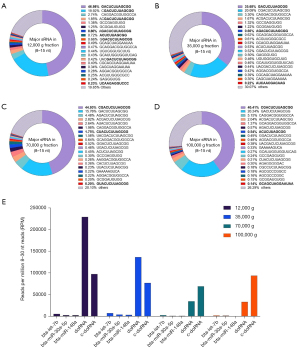
When looking specifically at these two doRNAs, 12K and 35K g fractions were more closely related to each other, with their most abundant sequence being the 12-nt doRNA (Figure 4A,4B). In subsequent fractions (70K and 100K g), the most abundant 8–15 sRNA was the 13-nt c-doRNA (Figure 4C,4D). These findings translated in a c-doRNA/doRNA ratio of 0.42, 0.56, 1.99 and 2.85 in 12K, 35K, 70K and 100K g fractions, respectively, which tended to increase with ultracentrifugation speeds (Figure 4A-4D). A closer inspection of the distribution profiles of c-doRNA and doRNA (Figure 4E) revealed that the c-doRNA/doRNA ratio is increasing mainly because doRNA sequences sediment at lower speeds, leaving less of these sequences for the subsequent fractions, while c-doRNA remains constant across all the fractions (Figure 4E). Therefore, these observations suggest a specific enrichment of doRNA in the lower speed fractions and potential association with the large exRNA-enriched mEVs it contains (6).
When compared to milk’s most enriched microRNAs (i.e., bta-let-7b, bta-miR-30a-5p and bta-miR-148a), doRNA and c-doRNA were systematically more abundant (Figure 4E). Twelve-nt doRNA was 16 to 194 times more enriched in these fractions than these three microRNAs, while 13-nt c-doRNA reached 12- to 553-fold higher abundance compared to bta-let-7b, bta-miR-30a-5p and bta-miR-148a (Figure 4E).
These sequencing results suggest that 12- and 13-nt doRNAs (I) are possibly more abundant than the most abundant milk microRNAs, (II) are specifically distributed across milk ultracentrifugation fractions, and (III) might be associated to the specific milk mEV subsets that sediment at the indicated speeds.
qPCR validation and comparison to the most enriched milk microRNAs
As sequencing data often trade large spectrum of detection for precision in quantification, we aimed to confirm these results using a complimentary and previously validated splint-ligation LNA-based RT-qPCR method designed to detect sRNA shorter than 15 nt (24).
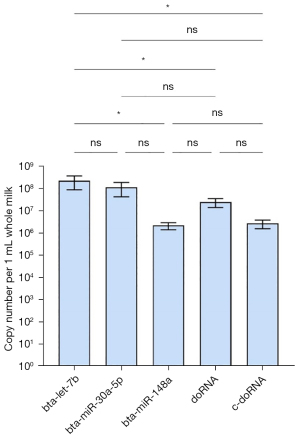
We first looked at the enrichment of doRNA and c-doRNA in unfractionated pasteurized milk, and compared those to the most enriched commercial milk microRNAs (16), namely bta-let-7a, bta-miR-30a-5p and bta-miR-148a (Figure 5). Both doRNA and c-doRNA were detectable in unfractionated commercial milk at a concentration of 106 to 107 copies per 1 mL milk (Figure 5). While doRNA and c-doRNA were slightly, yet not significantly, more enriched than bta-miR-148a, their levels were not higher than bta-miR-30a-5p and significantly lower than bta-let-7b (Figure 5).
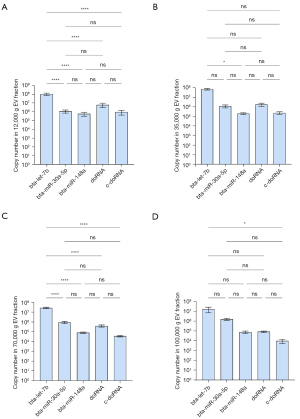
We observed similar results in milk fractions, with bta-let-7b systematically being the most enriched of the five small RNAs surveyed in all fractions (Figure 6). In the first two fractions (12K and 35K g), doRNA ranked second, followed by bta-miR-30a-5p, c-doRNA and, finally, bta-miR-148a (Figure 6A,6B). In the latter fractions, bta-miR-30a-5p ranked second, while doRNA was relocated to the third position and c-doRNA to the last, translating a difference in enrichment pattern between the fractions (Figure 6C,6D). When looking at distribution patterns across the fractions (Figure 7), distribution profiles of milk microRNAs matched our previous reports (6,16), with the bulk of bta-let-7b and bta-miR-148a significantly concentrating within the 12K and 35K g, while bta-miR-30a was distributed more evenly across the fractions (Figure 7A-7C). Similarly, doRNA and c-doRNA were more enriched within the first two milk fractions (Figure 7D,7E), confirming a differential enrichment of doRNA between milk fractions and associated mEVs.
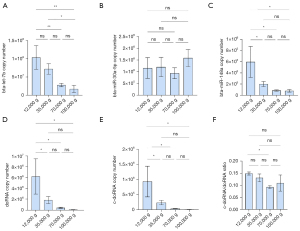
The trend of increasing c-doRNA/doRNA ratio with increasing ultracentrifugation speeds observed in our sRNA-seq data was not confirmed by RT-qPCR data (Figure 7F). There was, however, a significant difference in the c-doRNA/doRNA ratio between the 12K and 70K g fractions, supporting the potential specific loading of certain doRNAs within certain EV subsets (Figure 7F).
Discussion
Canonical sRNA sequencing pipelines rely on an arbitrary minimal length threshold of 16 nt for library preparation and subsequent bioinformatics analysis (21). This threshold was initially set to ensure higher depth of analysis for longer sRNA, improve the signal-to-noise ratio and normalize the downstream computational analyses. It was chosen on the premise that RNA species shorter than 15 nt may not be specific, might not be mapped with confidence to the genome and would not have biological significance, being most likely degradation products, artifacts or noise “polluting” the sequencing data. While better depth of analysis is indeed of importance in sRNA sequencing, such reasoning follows the same flawed paradigm of “junk short RNA” that hindered the discovery of microRNAs, piwi-interacting RNAs (piwiRNAs), tRFs and other small ribosomal RNAs (srRNAs) in the past (29,30).
The serendipitous discovery of 12-nt smiRNAs, as potentially competing with the function of the microRNA from which they derived, was among the first evidence questioning the relevance of that 16-nt threshold (20). Subsequently, we reported the discovery of other subsets of very-short RNAs, such as doRNAs, in animals (human cells and mouse cells and tissues), plants, yeasts and bacteria, further challenging this paradigm (19). Our experiments and controls confirmed that doRNAs were not mere processing artifacts, but rather biologically relevant and active short rRNAs implicated in cell proliferation and mRNA translation (19). By unveiling the existence of doRNAs in cow’s milk and fractions, along with a large array of RNAs shorter than the canonical sRNA-seq threshold of 15 nt, the evidence that we gathered here further supports this paradigm shift and extends the reach of doRNAs to the realm of exRNAs.
In addition, the focus on microRNAs in milk trended in the last decade because of their known important functions and because it was embedded within the larger “nutritional microRNAs” trend in research, leaving aside numerous biologically relevant exRNAs [reviewed in (1,11,28)]. However, mounting evidence suggests the existence of more than microRNAs in biological fluids, with several papers reporting the existence of other exRNAs, such as mRNAs, tRNAs, circRNAs and rRNAs in milk and mEVs (4,10,16,31-35). The results of the present study are in accordance with these reports and further emphasize on the importance of looking at the bigger picture rather than focusing on a single RNA species (36-38).
Interestingly, in this work, milk fractions and the mEVs they contain, were shown to be highly enriched in the 12- and 13-nt doRNAs, which constituted the vast majority of all 8–15 nt exRNAs, while this was not the case for the platelet EVs that we studied and reported in our previous work (19). Along with this observation, the distribution pattern of doRNAs and specific c-doRNA over doRNA ratios across milk fractions suggest that these 12- and 13-nt exRNAs might be specifically secreted within certain mEV subsets, as reported previously for tRNAs, microRNAs and iso-miRNAs (16,39-41). However, because of their length, further research is needed to clarify if these very small RNAs are bound by the same rules that guide other small RNAs with specific motives towards EVs or if it is their potential association with certain RNA-binding proteins that make doRNAs differentially enriched in milk fractions (19,42,43).
It was previously suggested that ultracentrifugation might be a degradative process (44,45). We cannot exclude the possibility that the observed impoverishment of doRNA within the last two fractions might be due to the loss of mEVs protecting these very-short RNAs from degradation by milk ribonucleases (46,47). Therefore, investigation of doRNAs in the different mEV subsets isolated through less disruptive methods (e.g., tangential filtration or size-exclusion chromatography) is warranted before drawing definitive conclusions about the specific loading of milk doRNAs within the larger mEVs found in the 12K and 35K g ultracentrifugation fractions (6). Moreover, as there is a possibility for exRNAs to be associated with more than simply EVs, including exosomes, there is a possibility that doRNAs in milk are associated with non-vesicular particles and ribonucleoproteins (48-50). Accordingly, further exploration of doRNA resistance to digestion and the exact mechanism of their secretion and transport in milk remains to be fully elucidated.
As exRNAs in milk have been previously found to resist digestion and potentially impact the health status of the “consumer”, be it cells, mice or humans (18,22,51,52), one might speculate that these new exRNAs in milk also contribute to the bioactivity of mEVs [reviewed in (8,10,28,53)]. In this line, we previously reported that doRNAs might be involved in prostate cancer progression, in which they are underexpressed in patient’s cells (19). These doRNAs also seemed to limit cell proliferation and regulation of the expression of AXIIR, which is an important receptor involved in the etiology of this disease (19). Overexpression of the doRNA-binding protein hnRNP A2/B1 in prostate cancer (54), its importance in the progression of this malignancy and for the resistance to apoptosis (55) would make of this protein a “hub”—and association with doRNAs (19)—potentially linking doRNAs and prostate cancer. While the link between milk consumption and prostate cancers remains controversial, with inconsistent evidence in the general population of men (56-58), milk and its bioactive components, including exRNAs, might affect the etiology of prostate cancer, more likely in those with digestive tracts more permeable to mEVs, as seen for bacterial lipopolysaccharide (LPS)-bearing EVs (59,60). This link, although speculative, calls for further investigations.
DoRNAs might also impact the maturation or degradation of rRNAs by binding to the 5’ end of 28S rRNA (1). This could interfere with the proper assembly and integrity of the nucleolus, affecting the accurate synthesis and processing of rRNAs and, if leading 28S rRNA to degradation or edition, potentially modulating the diversity of rRNAs (2). In turn, this could impact the function of ribosomes and disrupt mRNA translation. Therefore, doRNAs may have an impact on cell viability by modulating rRNA biogenesis (3).
To our knowledge, our study is the first to explore such very short exRNAs in milk, any other biological fluid and mEV-enriched ultracentrifugation fractions. Our sequencing results were supported by complementary high sensitivity, high specificity, LNA-based RT-qPCR, further confirming the existence of these extracellular doRNAs along with other very short exRNAs in commercial dairy cow’s milk.
While original, our work is, however, limited in several aspects. First, these experiments were conducted on a specific milk brand that is pasteurized and ultrafiltered. In accordance with previous reports [reviewed in (8,37,61,62)], milk processing for consumption highly impacts the exRNA content (63), with marked differences between ultra-high temperature (UHT) processing and pasteurized fluids (64). Even though we detected such very short exRNAs in other milks during our investigations and protocol developments (data not yet published), the results we report here might not be inferable to all cow milks around the globe, UHT milks, pascalized fluids (65) or milks from other species. Additionally, as for other exRNAs in milk, their transfer to recipient cells remains to be fully demonstrated, despite reports supporting this hypothesis (8,28,51).
In addition, because of their novelty, so far, the discovery and exploration of doRNAs in various tissues and conditions remains monocentric and requires replication from independent research groups worldwide, which we call for.
Slight modifications to any sRNA sequencing pipeline, aimed to allow for the detection of sequences shorter than 16 nt, might easily allow replication of these results and exploration of other very short exRNAs in other bodily fluids. Additional methods and information on how to explore these very small RNAs were also reported in our latest work and could serve as a guideline for exploring such small RNAs by sequencing across species, including bacteria, yeast and plants (43). However, one should exercise caution when analyzing sequencing data because of the discrepancies we report in this study between the sequencing data and quantitative analyses. Indeed, because classic RT-qPCR would not differentiate between these doRNA and 5.8S rRNA (or longer rRFs), we set, validated, and shared the protocol of a splint-ligation LNA-based RT-qPCR method which allows the highly specific, absolute quantification of doRNAs in any sample and under any condition of health and disease (24). Using this method, we confirmed that doRNAs were highly enriched in milk (at levels comparable to the most enriched milk microRNAs), but their concentration did not seem to be above that of microRNAs, as suggested by the initial sequencing analysis. Moreover, the important differences in doRNA/c-doRNA ratio observed in sequencing data were less pronounced when assessed by LNA RT-qPCR.
These differences might have risen from RNA isolation procedure necessarily different for each method. Indeed, while on-column isolation is more suitable for RNA sequencing, TRIzol LS is known to provide with different total RNA yields than columns (66), and both isolation techniques have been reported to have selectivity biases based on GC content (66-68). Also, our splint-ligation RT-qPCR method is reinforced with two internal spike-in controls for extraction and RT-qPCR efficiency estimation, two normalization tools that cannot be used for sequencing purposes. Therefore, while our sequencing analyses led to the discovery of doRNAs as a new kind of exRNAs, we believe our qPCR validation data to be more reliable. Our robust and easily applicable splint-ligation LNA-based RT-qPCR methodology may facilitate replication of our work and allow further exploration of doRNA and very short exRNA biology in different model organisms, biological fluids and contexts.
Conclusions
In this work, we report the discovery of very short exRNAs, most of which were rRNA-derived doRNAs, in commercially available cow’s milk and their specific distribution across EV-enriched ultracentrifugation fractions. The existence of such short exRNAs in milk further challenges arbitrary size paradigms in the molecular biology of sRNAs, which does not go without reminding of the micropeptide paradigm shift in the last decade (69-71). The existence of very short exRNA, such as doRNAs in milk, suggests their potential enrichment in other biological fluids, such as plasma, cell culture mediums, intercellular or amniotic fluid; a possibility to consider when exploring the involvement of exRNAs in physiological processes, pathological conditions or even for basic in vitro research (72). Further investigations are warranted to unveil the origin of milk doRNAs, their functions in different conditions, their potential as biomarkers of diseases, especially during lactation disorders and mammary gland infections, their transmission between cells, organs and possibly their bioactivity after transmission between individuals during breastfeeding or species through ingestion of cow’s milk.
Acknowledgments
Funding: This work was supported by the Fonds de recherche du Québec—Santé (Montréal, Canada), through PhD Studentship Award No. 262093 (to AB), the Fonds de Recherche du Québec—Nature et Technologies (Montréal, Canada), through PhD Studentship Award No. 289637 (to ML), and the Canadian Institutes of Health Research (CIHR, Ottawa, Canada) through grant number PJT-165806 (to PP).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, ExRNA for the series “Dietary MicroRNAs: Focus on Milk”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-6/rc
Data Sharing Statement: Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-6/dss
Peer Review File: Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-6/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-6/coif). The series “Dietary MicroRNAs: Focus on Milk” was commissioned by the editorial office without any funding or sponsorship. AB reports a doctoral studentship support from the Fonds de recherche du Québec-Santé. ML reports a doctoral studentship support from the FRQ-NT. PP served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of ExRNA. PP also reports an operating research grant support from the Canadian Institutes of Health Research (CIHR, Ottawa, Canada) paid to his Institution. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alsaweed M, Hartmann PE, Geddes DT, et al. MicroRNAs in Breastmilk and the Lactating Breast: Potential Immunoprotectors and Developmental Regulators for the Infant and the Mother. Int J Environ Res Public Health 2015;12:13981-4020. [Crossref] [PubMed]
- Lässer C, Shelke GV, Yeri A, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol 2017;14:58-72. [Crossref] [PubMed]
- Feng X, Chen X, Zheng X, et al. Latest Trend of Milk Derived Exosomes: Cargos, Functions, and Applications. Front Nutr 2021;8:747294. [Crossref] [PubMed]
- Tingö L, Ahlberg E, Johansson L, et al. Non-Coding RNAs in Human Breast Milk: A Systematic Review. Front Immunol 2021;12:725323. [Crossref] [PubMed]
- Kompaneets IY, Ermakov EA, Sedykh SE, et al. Secretory immunoglobulin A from human milk hydrolyzes microRNA. J Dairy Sci 2020;103:6782-97. [Crossref] [PubMed]
- Benmoussa A, Ly S, Shan ST, et al. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow's milk. J Extracell Vesicles 2017;6:1401897. [Crossref] [PubMed]
- Benmoussa A, Lee CH, Laffont B, et al. Commercial Dairy Cow Milk microRNAs Resist Digestion under Simulated Gastrointestinal Tract Conditions. J Nutr 2016;146:2206-15. [Crossref] [PubMed]
- Melnik BC, Stremmel W, Weiskirchen R, et al. Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 2021;11:851. [Crossref] [PubMed]
- Carrillo-Lozano E, Sebastián-Valles F, Knott-Torcal C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020;12:3066. [Crossref] [PubMed]
- Zeng B, Chen T, Luo JY, et al. Biological Characteristics and Roles of Noncoding RNAs in Milk-Derived Extracellular Vesicles. Adv Nutr 2021;12:1006-19. [Crossref] [PubMed]
- Melnik BC, Schmitz G. MicroRNAs: Milk's epigenetic regulators. Best Pract Res Clin Endocrinol Metab 2017;31:427-42. [Crossref] [PubMed]
- Leroux C, Chervet ML, German JB. Perspective: Milk microRNAs as Important Players in Infant Physiology and Development. Adv Nutr 2021;12:1625-35. [PubMed]
- Lawrie CH. editor. MicroRNAs in Medicine. Wiley, 2013.
- Benmoussa A, Michel S, Gilbert C, et al. Isolating Multiple Extracellular Vesicles Subsets, Including Exosomes and Membrane Vesicles, from Bovine Milk Using Sodium Citrate and Differential Ultracentrifugation. Bio Protoc 2020;10:e3636. [Crossref] [PubMed]
- Wang Y, Zhang Y, Kong L, et al. tRNA-derived RNA fragments in the exosomes of bovine milk and colostrum. Journal of Food Composition and Analysis 2021;102:103948. [Crossref]
- Benmoussa A, Laugier J, Beauparlant CJ, et al. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci 2020;103:16-29. [Crossref] [PubMed]
- Sukreet S, Braga CP, An TT, et al. Isolation of extracellular vesicles from byproducts of cheesemaking by tangential flow filtration yields heterogeneous fractions of nanoparticles. J Dairy Sci 2021;104:9478-93. [Crossref] [PubMed]
- Izumi H, Kosaka N, Shimizu T, et al. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012;95:4831-41. [PubMed]
- Lambert M, Benmoussa A, Diallo I, et al. Identification of Abundant and Functional dodecaRNAs (doRNAs) Derived from Ribosomal RNA. Int J Mol Sci 2021;22:9757. [Crossref] [PubMed]
- Plante I, Plé H, Landry P, et al. Modulation of microRNA Activity by Semi-microRNAs. Front Genet 2012;3:99. [Crossref] [PubMed]
- Dard-Dascot C, Naquin D, d'Aubenton-Carafa Y, et al. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 2018;19:118. [Crossref] [PubMed]
- Baier SR, Nguyen C, Xie F, et al. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014;144:1495-500. [Crossref] [PubMed]
- Benmoussa A, Gotti C, Bourassa S, et al. Identification of protein markers for extracellular vesicle (EV) subsets in cow's milk. J Proteomics 2019;192:78-88. [Crossref] [PubMed]
- Lambert M, Benmoussa A, Provost P. A New Specific and Sensitive RT-qPCR Method Based on Splinted 5' Ligation for the Quantitative Detection of RNA Species Shorter than microRNAs. Noncoding RNA 2021;7:59. [Crossref] [PubMed]
- Elsik CG, Unni DR, Diesh CM, et al. Bovine Genome Database: new tools for gleaning function from the Bos taurus genome. Nucleic Acids Res 2016;44:D834-9. [Crossref] [PubMed]
- Heintz-Buschart A, Yusuf D, Kaysen A, et al. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol 2018;16:52. [Crossref] [PubMed]
- Heberle H, Meirelles GV, da Silva FR, et al. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 2015;16:169. [Crossref] [PubMed]
- Benmoussa A, Provost P. Milk MicroRNAs in Health and Disease. Compr Rev Food Sci Food Saf 2019;18:703-22. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 2003;25:930-9. [Crossref] [PubMed]
- Ishikawa H, Rahman MM, Yamauchi M, et al. mRNA Profile in Milk Extracellular Vesicles from Bovine Leukemia Virus-Infected Cattle. Viruses 2020;12:669. [Crossref] [PubMed]
- Zhou Y, Yu Z, Wang X, et al. Exosomal circRNAs contribute to intestinal development via the VEGF signalling pathway in human term and preterm colostrum. Aging (Albany NY) 2021;13:11218-33. [Crossref] [PubMed]
- Karlsson O, Rodosthenous RS, Jara C, et al. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: Implications for early child development. Epigenetics 2016;11:721-9. [Crossref] [PubMed]
- Zeng B, Chen T, Luo J, et al. Exploration of Long Non-coding RNAs and Circular RNAs in Porcine Milk Exosomes. Front Genet 2020;11:652. [Crossref] [PubMed]
- Zeng B, Chen T, Xie MY, et al. Exploration of long noncoding RNA in bovine milk exosomes and their stability during digestion in vitro. J Dairy Sci 2019;102:6726-37. [Crossref] [PubMed]
- Zempleni J. Milk exosomes: beyond dietary microRNAs. Genes Nutr 2017;12:12. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Exosomes of pasteurized milk: potential pathogens of Western diseases. J Transl Med 2019;17:3. [Crossref] [PubMed]
- Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J 2013;12:103. [Crossref] [PubMed]
- Nolte-'t Hoen EN, Buermans HP, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res 2012;40:9272-85. [Crossref] [PubMed]
- Corrado C, Barreca MM, Zichittella C, et al. Molecular Mediators of RNA Loading into Extracellular Vesicles. Cells 2021;10:3355. [Crossref] [PubMed]
- Wozniak AL, Adams A, King KE, et al. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J Cell Biol 2020;219:e201912074. [Crossref] [PubMed]
- Garcia-Martin R, Wang G, Brandão BB, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022;601:446-51. [Crossref] [PubMed]
- Lambert M, Guellal S, Ho J, et al. An Expanded Landscape of Unusually Short RNAs in 11 Samples from Six Eukaryotic Organisms. Noncoding RNA 2022;8:34. [Crossref] [PubMed]
- Lamparski HG, Metha-Damani A, Yao JY, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods 2002;270:211-26. [Crossref] [PubMed]
- Tzaridis T, Bachurski D, Liu S, et al. Extracellular Vesicle Separation Techniques Impact Results from Human Blood Samples: Considerations for Diagnostic Applications. Int J Mol Sci 2021;22:9211. [Crossref] [PubMed]
- Soboleva SE, Sedykh SE, Alinovskaya LI, et al. Cow Milk Lactoferrin Possesses Several Catalytic Activities. Biomolecules 2019;9:208. [Crossref] [PubMed]
- Nevinsky GA, Zakharova OD, Kompaneets IY, et al. Six catalytic activities and cytotoxicity of immunoglobulin G and secretory immunoglobulin A from human milk. J Dairy Sci 2021;104:6431-48. [Crossref] [PubMed]
- Umu SU, Langseth H, Bucher-Johannessen C, et al. A comprehensive profile of circulating RNAs in human serum. RNA Biol 2018;15:242-50. [Crossref] [PubMed]
- Geekiyanage H, Rayatpisheh S, Wohlschlegel JA, et al. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc Natl Acad Sci U S A 2020;117:24213-23. [Crossref] [PubMed]
- Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol 2012;9:1066-75. [Crossref] [PubMed]
- Mutai E, Ramer-Tait AE, Zempleni J. MicroRNAs in bovine milk exosomes are bioavailable in humans but do not elicit a robust pro-inflammatory cytokine response. ExRNA 2020;2:2. [Crossref]
- Wiklander OP, Nordin JZ, O'Loughlin A, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles 2015;4:26316. [Crossref] [PubMed]
- Jiang X, You L, Zhang Z, et al. Biological Properties of Milk-Derived Extracellular Vesicles and Their Physiological Functions in Infant. Front Cell Dev Biol 2021;9:693534. [Crossref] [PubMed]
- Stockley J, Villasevil ME, Nixon C, et al. The RNA-binding protein hnRNPA2 regulates β-catenin protein expression and is overexpressed in prostate cancer. RNA Biol 2014;11:755-65. [Crossref] [PubMed]
- Zhang H, Chen N, Wang X, et al. Identification and validation of hub genes in prostate cancer progression based on weighted gene co-expression network analysis. Nan Fang Yi Ke Da Xue Xue Bao 2021;41:1631-40. [PubMed]
- Preble I, Zhang Z, Kopp R, et al. Dairy Product Consumption and Prostate Cancer Risk in the United States. Nutrients 2019;11:1615. [Crossref] [PubMed]
- López-Plaza B, Bermejo LM, Santurino C, et al. Milk and Dairy Product Consumption and Prostate Cancer Risk and Mortality: An Overview of Systematic Reviews and Meta-analyses. Adv Nutr 2019;10:S212-23. [Crossref] [PubMed]
- Sargsyan A, Dubasi HB. Milk Consumption and Prostate Cancer: A Systematic Review. World J Mens Health 2021;39:419-28. [Crossref] [PubMed]
- Melnik BC, John SM, Weiskirchen R, et al. The endocrine and epigenetic impact of persistent cow milk consumption on prostate carcinogenesis. J Transl Genet Genom 2022;6:1-45. [Crossref]
- Tulkens J, Vergauwen G, Van Deun J, et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 2020;69:191-3. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Pasteurized non-fermented cow's milk but not fermented milk is a promoter of mTORC1-driven aging and increased mortality. Ageing Res Rev 2021;67:101270. [Crossref] [PubMed]
- Kleinjan M, van Herwijnen MJ, Libregts SF, et al. Regular Industrial Processing of Bovine Milk Impacts the Integrity and Molecular Composition of Extracellular Vesicles. J Nutr 2021;151:1416-25. [Crossref] [PubMed]
- Howard KM, Jati Kusuma R, Baier SR, et al. Loss of miRNAs during processing and storage of cow's (Bos taurus) milk. J Agric Food Chem 2015;63:588-92. [Crossref] [PubMed]
- Zhang Y, Xu Q, Hou J, et al. Loss of bioactive microRNAs in cow's milk by ultra-high-temperature treatment but not by pasteurization treatment. J Sci Food Agric 2022;102:2676-85. [Crossref] [PubMed]
- Chughtai MFJ, Farooq MA, Ashfaq SA, et al. Role of Pascalization in Milk Processing and Preservation: A Potential Alternative towards Sustainable Food Processing. Photonics 2021;8:498. [Crossref]
- Oh S, Park MR, Son SJ, et al. Comparison of Total RNA Isolation Methods for Analysis of Immune-Related microRNAs in Market Milks. Korean J Food Sci Anim Resour 2015;35:459-65. [Crossref] [PubMed]
- Mateescu B, Kowal EJ, van Balkom BW, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles 2017;6:1286095. [Crossref] [PubMed]
- McAlexander MA, Phillips MJ, Witwer KW. Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid. Front Genet 2013;4:83. [Crossref] [PubMed]
- Lee C, Zeng J, Drew BG, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 2015;21:443-54. [Crossref] [PubMed]
- Yeasmin F, Yada T, Akimitsu N. Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Front Genet 2018;9:144. [Crossref] [PubMed]
- Tharakan R, Sawa A. Minireview: Novel Micropeptide Discovery by Proteomics and Deep Sequencing Methods. Front Genet 2021;12:651485. [Crossref] [PubMed]
- Mannerström B, Paananen RO, Abu-Shahba AG, et al. Extracellular small non-coding RNA contaminants in fetal bovine serum and serum-free media. Sci Rep 2019;9:5538. [Crossref] [PubMed]
Cite this article as: Benmoussa A, Husseini Z, Ho J, Guellal S, Lambert M, Gilbert C, Provost P. DodecaRNAs (doRNAs) are abundant in cow’s milk and differentially enriched in milk ultracentrifugation fractions. ExRNA 2022;4:20.


