
miRNAs provide mechanisms for integrated control of endocrine pancreas homeostasis and metabolic disease pathogenesis: a narrative review
Introduction
In a healthy individual, insulin secretion accurately matches the insulin demand of target tissues such as skeletal muscle and white adipose tissue, therefore maintaining glucose homeostasis. However, in models of insulin resistance or type 2 diabetes (T2D), compensatory responses to increase insulin secretion and/or expand beta-cell mass occur until beta-cell failure ensues (1,2). The initial adaptive responses are part of allostatic mechanisms to maintain adequate glucose homeostasis, whereas the latest is a deleterious end-point characterised by insufficient/inadequate insulin secretion and beta-cell destruction. These allostatic adaptations to maintain energy homeostasis integrate beta-cell mass and associated insulin levels and secretion with the function of other organs. The adaptation at the level of the pancreatic islets is controlled by multiple biological processes coordinating proliferation, differentiation, and ultimately the death of beta-cells, involving the regulation of insulin gene expression, protein translation, and the mechanisms responsible for insulin secretion (3,4). Our understanding of endocrine pancreas pathobiology in T2D and T1D has been enriched by the contribution of miRNAs to these diseases.
microRNAs (miRNAs) are endogenous short non-coding RNAs, usually containing 19–22 nucleotides, that repress translation or initiate mRNA degradation through binding (mostly) to the 3'UTR of mRNAs (5,6). During the last decades, miRNAs have emerged as novel and regulatory effectors on gene transcription at the intracellular level. Moreover, the origin of these miRNAs might be in distant organs from where they are transported as targeted cargo in exosomes (7). The discovery of miRNAs and their “endocrine/paracrine” role has opened exciting new avenues for metabolic research aimed at understanding how and where they originated, which genes they target, when that occurs, and what (patho)physiological outcomes are associated with their dysregulation.
The current interest in miRNAs in health and disease has been fuelled by technologies such as genetic engineering and advanced analytical tools to dissect their contributions to physiological and pathophysiological outcomes (8,9). A compelling example of miRNA network dysregulation in diabetes comes from the research by Melkman-Zehavi and colleagues that showed how a beta-cell-specific deletion of Dicer1 in vivo prevents pre-miRNAs’ cleavage, leading to impaired insulin secretion and recapitulating the natural progression of diabetes (10).
The study of miRNAs in metabolism is novel and promising but not without significant challenges and unsolved questions: (I) the majority of studies focus on the role or association of individual miRNA with the onset or natural history of the disease; (II) there is evidence that a unique miRNA can target multiple targets genes (as a coordinated effect) or conversely, multiple miRNAs can target the same gene limiting our understanding of the real-size effect of miRNAs and the synergies among them in the pathogenesis of T2D; (III) there is still limited information about the transcriptional regulation of the miRNA; and (IV) lack of integrative approaches to investigate the crosstalk between different organs using miRNAs as proxy information molecule at the organismal level. Some of these limitations have been recently addressed by several comprehensive investigations (I) using data from T2D cohorts and miRNA-mRNA network-based approaches, confirming some classical and newly identified miRNAs linked to T2D and identifying clusters of target genes associated with functional aspects of islets and insulin secretion (11) and (II) taking advantage of machine learning workflows and identifying specific miRNA signatures associated with insulin transcription (12).
In this review, we focus on the roles of miRNAs in pancreas endocrine biology and integrate the knowledge of well-established and recent discoveries using genetically modified organisms (mostly mouse models) and in vitro models. We summarise how miRNAs affect islet function in the context of insulin resistance and T2D (and also, to a lesser extent, T1D) with a focus on functional roles in beta-cell proliferation, differentiation, apoptosis, insulin biosynthesis and secretion (Table 1).
Table 1
| miRNAs ID | Expression profile | Phenotype | Target(s) | Model(s) | References |
|---|---|---|---|---|---|
| miR-101a | Increased by inflammatory cytokines | Increases β-cell apoptosis; decreases GSIS; decreases insulin biosynthesis | NeuroD1, Onecut2 | MIN6 (mouse) | (13) |
| miR-124a | Increased in T2D patients | Decreases insulin biosynthesis; decreases GSIS | NeuroD1, Mtpn, Foxa2 | MIN6 (mouse) | (14,15) |
| miR-125b | Increased by high glucose | Decreases insulin biosynthesis; decreases GSIS | M6pr, Mtfp1 | MIN6 (mouse) human islets; mouse | (16) |
| miR-126 | Increased by high glucose | Decreases β-cell proliferation | Irs2 | INS-1 (rat) | (17) |
| miR-127 | Decreased in obese mice; increased by high glucose | Decreases β-cell proliferation; decreases GSIS | Kif3b | MIN6 (mouse) | (18) |
| miR-130 | Increased in T2D patients and rats | Decreases insulin biosynthesis; decreases GSIS | INS-1 (rat) | (19) | |
| miR-132 | Increased in T2D mice | Increases GSIS | INS-1 (rat)E; mouse | (20,21) | |
| miR-133a | Increased in T2D patients | Decreases insulin biosynthesis | Ptbp | Human islets | (22,23) |
| miR-139-5p | Decreases GSIS | Pick1 | MIN6 (mouse); mouse | (24) | |
| miR-141 | Increased in diabetic mice and human | Decreases β-cell proliferation; decreases GSIS | Foxa2 | INS-1 (rat), MIN6 (mouse) | (25) |
| miR-145 | Decreased in T2D rat | Increases β-cell apoptosis; decreases β-cell differentiation; decreases GSIS | Xbp1, Sox2, Abca1 | MIN6 (mouse); HuAEC-derived β islet-like cells mouse islets; rat | (26-28) |
| miR-146 | Increased in T2D patients; increased in obese mice; increased by high glucose | Increases β-cell apoptosis | MIN6 (mouse); rat islets | (29,30) | |
| miR-149-5p | Decreased by high glucose | Decreases β-cell apoptosis; increases GSIS | Bim | MIN6 (mouse) | (31) |
| miR-15a | Increases insulin biosynthesis; increases GSIS | Ucp2 | MIN6 (mouse) | (32) | |
| miR-152 | Decreased in T2D patients | Increases β-cell proliferation; increases GSIS | PI3Kα, Pdha1 | INS-1 (rat) MIN6 (mouse) | (19,33) |
| miR-153 | Increased in T2D patients and mice | Decreases GSIS | Vamp2, Snap25, Stx1a, Cacna1c | INS-1 (rat) MIN6 (mouse); mouse islets; mouse | (34,35) |
| miR-155-5p | Increased by hyperlipidemia in mice | Increases insulin biosynthesis | Mafb | MIN6 (mouse); mouse | (36) |
| miR-17 | Increased by high glucose | Increases β-cell proliferation | Menin | MIN6 (mouse) | (37) |
| miR-17-92 | Increased in obese mice; increased by high glucose | Increases β-cell proliferation; decreases GSIS; decreases insulin biosynthesis | NeuroD1 | MIN6 (mouse); mouse | (38,39) |
| miR-181a-5p | Increases β-cell apoptosis | INS-1 (rat) | (40) | ||
| miR-181c-5p | Increases β-cell differentiation | Smad7, Tgif2 | hiPSC | (41) | |
| miR-184 | Decreased in T2D patients and mice | Decreases β-cell proliferation; decreases GSIS | Ago2 | MIN6 (mouse) | (42) |
| miR-185 | Decreased in T2D patients and mice | Increases β-cell proliferation; increases GSIS; decreases β-cell apoptosis | Socs3 | MIN6 (mouse) | (43) |
| miR-187 | Increased in T2D patients | Decreases GSIS | Hipk3 | INS-1 (rat)at islets | (44) |
| miR-19a-3p | Increased in T2D patients | Increases β-cell proliferation; increases GSIS | Socs3 | INS-1 (rat) MIN6 (mouse) | (45) |
| miR-190b | Decreases β-cell proliferation; decreases GSIS | Nkx6.1 | MIN6 (mouse) | (46) | |
| miR-196b | Increased by high glucose | Increases insulin biosynthesis | insulin 2 | βTC6 (mouse) | (47) |
| miR-199b-5p | Increased in pancreas regeneration rodent model | Increases β-cell proliferation | Mlk3 | Rat islets | (48) |
| miR-200 family | Increased in T2D patients and mice | Increases β-cell apoptosis; decreases GSIS | Dnajc3, Xiap, Jazf1, Ypel2, Rps6kb1, Etv5 | MIN6 (mouse); INS-1E (rat) EndoC-βH1 (human); human islets; mouse | (49,50) |
| miR-204 | Increased in obese mice | Decreases insulin biosynthesis | Mafa (controversial) | INS-1 (rat) EndoC-βH1 (human); human islets; mouse islets | (51,52) |
| miR-205-5p | Increased in T2D mice | Increases insulin biosynthesis | Tcf7l2 | INS-1 (rat) | (53) |
| miR-21 | Increased in T2D mice | Decreases β-cell differentiation; increases GSIS | Sox6, Rbpj | Embryonic pancreas (chick); mouse | (54,55) |
| miR-212 | Increases GSIS | Crct1 | INS-1 (rat); mouse | (56) | |
| miR-217 | Increased in T2D patients and mice | Decreases β-cell proliferation; increases β-cell apoptosis | Mafb | β-TC-tet (mouse) | (57) |
| miR-218 | Decreased by high glucose | Decreases GSIS | Stxbp1 | MIN6 (mouse); mouse islets | (58) |
| miR-221/222 | Increased in obese mice | Increases β-cell proliferation; decreases β-cell apoptosis; decreases insulin biosynthesis | Nfatc3, Pak1 | MIN6 (mouse); INS-1 (rat); mouse | (59,60) |
| miR-223 | Increased in obese and diabetic mice and human | Increases β-cell proliferation; increases GSIS | Foxo, Sox6 | MIN6 (mouse); mouse | (61) |
| miR-23 | Decreased by inflammation | Decreases β-cell apoptosis | EndoC-βH1 (human) | (62) | |
| miR-24 | Increased in obese mice | Decreases β-cell proliferation; decreases GSIS | Sp1, Hnf1α, NeuroD1 | MIN6 (mouse); mouse islets | (63,64) |
| miR-25 | Increased in T2D mice | Decreases insulin biosynthesis | Ins1 | INS-1 (rat) | (65) |
| miR-26 | Increases β-cell differentiation | Tet1/2/3, Tdg | Mouse | (66) | |
| miR-27a | Increased in obese mice | Increases β-cell apoptosis | INS-1 (rat) | (67) | |
| miR-29a | Increased by high glucose | Decreases GSIS | Stx1a | INS-1E (rat) | (68) |
| miR-296-3p | Decreased in diabetic; patients | Decreases β-cell apoptosis | Pten | MIN6 (mouse) | (69) |
| miR-297b-5p | Decreased by stearic acid in mice | Decreases β-cell apoptosis | Lats2 | β-TC6 (mouse); mouse islets | (70) |
| miR-299-5p | Decreased by glucolipotoxicity in primary human islets | Decreases β-cell apoptosis | Perp, Siah1, Rassf6, Sgk1 | β-TC6 (mouse); MIN6 (mouse); mouse islets; human islets | (71) |
| miR-30a-5p | Increased in obese mice; increased by high glucose | Increases insulin biosynthesis | NeuroD1 | INS-1 (rat); mouse | (72) |
| miR-30b | Increased by inflammatory cytokines | Increases β-cell apoptosis; decreases insulin biosynthesis | Bcl2, NeuroD1 | MIN6 (mouse) | (13) |
| miR-30d | Decreased in T2D patients and mice | Decreases β-cell proliferation; increases β-cell apoptosis; increases insulin biosynthesis | Map4k4 | MIN6 (mouse); mouse islets; mouse | (73-75) |
| miR-32 | Increases β-cell differentiation | Wwp2 | (76) | ||
| miR-322 | Decreased by high glucose | Decreases GSIS | Stxbp1 | MIN6 (mouse); mouse islets | (58) |
| miR-335-5p | Increased in T2D patients and rat | Decreases β-cell proliferation; increases β-cell apoptosis; decreases GSIS | Glut-4, Snap25, Stxbp1, Syt11 | MIN6 (mouse); INS-1 (rat) EndoC-βH1 (human) | (77,78) |
| miR-338-3p | Decreased in obese and diabetic mice | Decreases β-cell proliferation; increases β-cell apoptosis | INS832/13 (rat); rat islets human islets; mouse | (79,80) | |
| miR-34a | Increased in T2D patients and mice | Increases β-cell apoptosis; decreases GSIS | Vamp2, Bcl2, Sirt1 | INS-1 (rat) | (81) |
| miR-344-5p | Decreased by lipotoxicity | Decreases β-cell apoptosis | Cav1 | INS-1 (rat) | (82) |
| miR-375 | Increased in T2D patients; increased obese mice | Decreases β-cell proliferation; increases β-cell apoptosis; increases β-cell differentiation; decreases GSIS; decreases insulin biosynthesis | Psen1, Mtpn, Yap1, Pdk1, Hnf1βb, Pax6, Insm1, Gata6, Pdpk1, Insr | NIT-1 (mouse) MIN6 (mouse); INS-1 (rat); mouse islets; rat islet; human islets; mouse | (83-90) |
| miR-383 | Decreased by high glucose | Decreases β-cell apoptosis | Tlr4, ApoC3 | MIN6 (mouse); mouse | (91) |
| miR-409-3p | Decreases β-cell apoptosis; increases GSIS | Casp8 | NES2Y (human) | (92) | |
| miR-433 | Decreased by high glucose | Increases β-cell proliferation; decreases β-cell apoptosis | Cox2 | MIN6 (mouse) | (93) |
| miR-455 | Increased in obese mice | Increases β-cell proliferation | Cpeb1 | MIN6 (mouse); EndoC-βH1 (human); human islets; mouse islets; mouse | (94) |
| miR-463-3p | Increased in T2D patients | Decreases GSIS | Abcg4 | MIN6 (mouse); mouse islets; human islets | (95) |
| miR-483 | Increased in T2D mice; increased by high glucose | Increases insulin biosynthesis; decreases β-cell apoptosis | Socs3, Aldh1a3 | βTC3 (mouse) MIN6 (mouse); mouse islets; mouse | (96,97) |
| miR-532-5p | Decreased by high glucose | Decreases β-cell apoptosis; increases GSIS | Ccnd1 | MIN6 (mouse) | (98) |
| miR-552-3p | Decreased in T2D patients | Decreases β-cell apoptosis | Jak1 | INS-1 (rat) | (99) |
| miR-577 | Increased in T1D patients | Decreases insulin biosynthesis; decreases GSIS | Fgf21 | INS-1 (rat) Embryonic pancreatic beta-cells | (100) |
| miR-7 | Increases β-cell differentiation | Gata6, Irs-2, Irs-1, Pax6, Pax4, NeuroD1, Nkx2.2 | hiPSC | (101) | |
| miR-7a | Decreased in T2D patients and mice | Decreases β-cell proliferation | p70S6K, eIF4E, Mknk1, Mknk2, Mapkap1 … | MIN6 (mouse); mouse islets human islets; mouse | (102-104) |
| miR-708 | Increased in obese mice | Increases β-cell apoptosis; decreases GSIS | Nnat | MIN6 (mouse); mouse islets | (105) |
| miR-802 | Increased in obese mice | Decreases GSIS; decreases insulin biosynthesis | NeuroD1, Fzd5 | MIN6 (mouse); mouse islets; Mouse | (106) |
| miR-9 | Decreases GSIS | Sirt1, Oc2, Stxbp1 | MIN6 (mouse); INS-1 (rat) | (107,108) | |
| miR-92a | Increased in T2D mice; decreased by high glucose | Decreases β-cell apoptosis; decreases insulin biosynthesis; increases GSIS | Klf2, Ins1 | MIN6 (mouse); INS-1 (rat) | (65,109) |
| miR-96 | Increased in T2D patients and mice | Increases β-cell proliferation; decreases β-cell apoptosis | Foxo1, Sox6 | MIN6 (mouse); mouse | (110) |
| Let7b-5p | Increased in T2D patients | Decreases β-cell proliferation; increases insulin biosynthesis; decreases GSIS | Ccnd1, Ccnd2 | INS-1 (rat); mouse | (111,112) |
miRNA, microRNA; T2D, type 2 diabetes; GSIS, glucose-stimulated insulin secretion; hiPSC, human induced pluripotent stem cell; INS, insulin gene.
The role of miRNAs on pancreatic cancer or other pancreatic pathologies falls out of the remit of this review and will not be addressed here. We remit the readers to previous excellent reviews on that subject (113-115). We present the following article in accordance with the Narrative Review reporting checklist (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-17/rc).
Methods
We used the PubMed database and restricted our search to manuscripts written in English until June 2022. We used general Boolean expressions as follows: (I) (microRNA OR miRNA OR miR) AND (pancreas OR islet OR beta cell); (II) (microRNA OR miRNA OR miR) AND pancreas AND crosstalk. Then, we applied several exclusion criteria (manually curated), such as (I) manuscripts unrelated to the specific topic of the review; (II) manuscripts related to tumours or cancer-related processes; (III) manuscripts that lacked novelty based on the authors’ discretion. Authors independently selected and ranked manuscripts based on their originality and the robustness of their findings, and a final selection of manuscripts was agreed. Several manuscripts were added to the review following reviewers’ suggestions. Our research strategy is outlined in Table 2.
Table 2
| Items | Specification |
|---|---|
| Date of search | May to June 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | (I) (microRNA OR miRNA OR miR) AND (pancreas OR islet OR beta cell); |
| (II) (microRNA OR miRNA OR miR) AND pancreas AND crosstalk | |
| Timeframe | Manuscripts published before June 2022 |
| Inclusion and exclusion criteria | Inclusion criteria: |
| (I) English only; | |
| (II) research manuscripts; | |
| (III) review manuscripts were only mentioned when referring to an area/field not addressed in the present review | |
| Exclusion criteria: | |
| (I) manuscripts unrelated to the specific topic of the review; | |
| (II) manuscripts related to tumours or cancer related processes; | |
| (III) manuscripts that lacked novelty based on authors’ discretion | |
| Selection process | Authors independently selected and ranked manuscripts based on their originality and the robustness of their findings and a final selection of manuscripts was agreed |
Evidence that miRNAs are essential for beta-cell proliferation
One of the first pieces of evidence of the role of miRNAs in pancreas biology came from studies showing that miRNAs target the expression of genes regulating beta-cell proliferation affecting insulin levels (83). The number of targeted genes, the regulation effect of the diabetic/glycemic status on the expression of a particular miRNA, and the outcome promoting or repressing beta-cell proliferation are surprisingly diverse (Figure 1). Globally considered, these studies evidence the complex regulation of miRNAs. Some of these miRNAs seem to operate as part of homeostatic responses aimed at maintaining appropriate levels of insulin and insulin secretion in the context of obese and insulin-resistant states by controlling beta-cell proliferation and beta-cell mass where the dysregulation of others a priori could worsen and exacerbate the T2D phenotype.
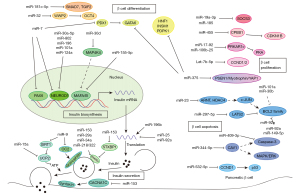
Several examples of miRNAs induced in T2D can be considered homeostatic regulatory events aimed at restoring glycemic levels. One of these miRNAs, described to exert positive effects on beta-cell proliferation, is miR-455, whose expression is induced in obese and diabetic models and mediated by the activation of Erg2. miR-455 inhibits the expression of Cpeb1 by directly targeting its 3'UTR. The decreased Cpeb1 suppresses the elongation of the poly(A) tail and the subsequent Xdkn1b translation, removing the inhibition of G1-S transition and promoting beta-cell proliferation (94). Another homeostatic miRNA upregulated by a high concentration of glucose is miR-17. Here, beta-cells overexpressing miR-17 proliferate driven by the inhibitory effect of miR-17 on Menin (a tumour suppressor) expression and the concomitant reduction of the downstream factors p21 and p27 (37). Following a similar regulatory pattern, miR-19a-3p is upregulated in the plasma of T2D patients, inducing the downregulation of Socs3—a negative regulator of cytokine signalling—via binding its 3'UTR enhancing beta-cell proliferation and insulin secretion (45). Other examples of miRNAs induced in T2D to promote beta-cell proliferation are miR-199b-5p and miR-223. miR-199b-5p directly interacts with Mlk3 and downregulates its expression (48). On the other hand, the knockout mouse of miR-223 shows impaired beta-cell proliferation and insulin secretion in vitro and glucose intolerance, but interestingly also, insulin resistance in vivo. The ablation of miR-223 triggers the activation of Foxo1 and Sox6 signalling cascades, which regulate the expression of beta-cell markers (Pdx1, Nkx6.1, Ucn3), and cell cycle-related genes (cyclin D1, cyclin E1, and P27), suggesting that miR-223 is a critical factor for maintaining functional beta-cell mass and adaptation during metabolic stress (61).
Interestingly, some examples of miRNA are also upregulated in hyperglycemic/T2D conditions that conversely impair beta-cell proliferation and contribute negatively to the pathogenesis of T2D. This is the case for miR-126, whose expression is upregulated in response to high glucose and insulin and suppresses beta-cell proliferation by directly targeting insulin receptor substrate 2 (Irs-2) (17). It is also worth mentioning the miR-190b, which impairs beta-cell proliferation and insulin secretion by targeting Nkx6.1, a mechanism that could mediate gestational diabetes mellitus (46). Similar outcomes have been described for miR-141. miR-141 is upregulated in T2D patients’ serum and mice islets. In vitro overexpression of miR-141 in rat insulinoma cells (INS-1) beta-cells leads to impaired cell proliferation and reduced glucose-stimulated insulin secretion (GSIS) by targeting the 3'UTR of Foxa2 transcript (25).
Other examples of how inappropriate levels of miRNA cause harmful effects on beta-cell proliferation come from models such as transgenic mice. This is the case of Let7b-5p, which is upregulated in the serum of T2D patients (111). At the cellular level, Let7b-5p-TG mice present a decreased expression of the proliferation marker Ki67, with smaller beta-cells and beta-cell mass, by regulating Ccnd1 and Ccnd2 mRNA (112).
Similarly, miR-17-92 is highly expressed in pancreatic islets and beta-cell lines, and its expression is upregulated in vivo in response to high-fat diet (HFD) and in vitro in response to high glucose. The beta-cell-specific knockout (KO) mice for miR-17-92 show a decreased beta-cell number and increased beta-cell size compared to the wildtype (WT) mice. In line with these data, the KO mice exhibit impaired glucose tolerance with a reduced insulin level. At the gene expression level, the KO mice present an upregulation of cell proliferation (Pten), cell cycle (Cdk1a, Rbl2) and transforming growth factor (TGF)-beta signalling (Tgfbr2, Smad2, Smad4) related genes, as well as the expression of several inhibitors of insulin production such as Sox6 and Crem likely as an attempt to compensate for the primary dysfunction caused by the genetic ablation of miR-17-92 (38).
On another note, several miRNAs exert beneficial effects whose expression is compromised in T2D, potentially contributing to its pathogenesis. For instance, miR-185 is downregulated in the islets of T2D patients and mice. Overexpression of miR-185 in beta-cells enhances cell proliferation and insulin secretion and protects beta-cells from apoptosis by directly targeting 3'UTR of Socs3 mRNA and positively regulates the STAT-3 signalling pathway (43). Another miRNA physiologically promoting beta-cell proliferation is miR-152, whose expression is negatively correlated with blood glucose levels in T2D patients. Its overexpression promotes beta-cell proliferation and insulin secretion by directly targeting 3'UTR of the PI3K catalytic subunit α (PI3Kα) and inhibiting its expression (33).
These results contrast with the adverse effects exerted by other miRNAs that contribute negatively to the pathogenesis of T2D and whose expression is downregulated in diabetes, such as miR-127. miR-127 is highly expressed in mouse beta-cells and downregulated in islets of mice fed with HFD and in beta-cell lines treated with high glucose concentrations. Interestingly, its overexpression attenuates beta-cell proliferation and inhibits insulin secretion by directly targeting and downregulating the expression of Kif3b (a kinesin family member) while having no impact on beta-cell apoptosis (18). miR-184, whose expression is also downregulated in islets of obese mice and T2D patients, directly targets Ago2, inhibiting the compensation of beta-cell proliferation and insulin secretion during insulin resistance (42). miR-7a is enriched in mouse beta-cells and directly targets some components of mTOR signalling pathway. Thus, inhibition of miR-7a activates mTOR signalling, including TORC1 and its downstream effectors p70S6 and eIF4E, and two MAPK-interacting kinases Mknk1 and Mknk2, but also Pax6, promoting beta-cell replication in mouse islets, which the treatment with rapamycin can reverse. These data evidence that miR-7a acts as a brake on adult beta-cell proliferation (103).
Another exciting example is miR-338-3p, whose expression is decreased in islets of obese and insulin-resistant mice. Interestingly, the use of miRNA sponges to inhibit the activity of this miRNA results in increased beta-cell proliferation without any significant change in beta-cell mass and GSIS. Curiously, its hosting gene, apoptosis-associated tyrosine kinase (AATK) has a similar impact as miR-338-3p on beta-cells (80).
miRNAs as gatekeepers for beta-cell apoptosis
An alternative strategy to maintain beta-cell mass is to prevent beta-cell apoptotic death. In the context of obesity, diabetes and a high-fat diet, we find associated changes in miRNAs in blood and islets. Their functional interpretation is complex since they might represent allostatic responses trying to maintain metabolic homeostasis or disease effectors. Moreover, these functions might be accomplished through increased levels and/or repression, and the analysis of the targets of these miRNAs also reveals heterogeneous mechanisms leading to similar pro/anti-apoptotic effects (Figure 1).
Several miRNAs protect against beta-cell apoptosis by directly or indirectly regulating Bcl2 family members and/or other independent targets. For instance, the miR-23 family, miR-23a-3p or miR-23b-3p are downregulated in beta-cells in response to pro-inflammatory cytokines. Their physiological role is to protect human beta-cells from inflammatory cytokine-induced cell death by downregulating the expression of pro-apoptotic Bcl2 family members, including Dp5, Puma, Bax and Bim, through a mechanism dependent on the activation of c-JUN pathway (62). Also, miR-92a and miR-149-5p inhibit cell apoptosis and reactive oxygen species (ROS) release and increase insulin secretion when beta-cell cultured with high glucose via directly targeting Klf2 and BCL2 like 11 (Bim), a BCL-2 protein family member, respectively. Both, miR-92a and miR-149-5p decrease their levels in beta-cells treated with high glucose (31,109). Another apoptosis protector is miR-297b-5p, which targets Lats2, a novel tumour suppressor reported to promote Bcl2 degradation (70), which is recently described as reducing the expression of the serum amyloid A3 (Saa3) (116). Also what relevant is that miR-297b-5p expression is downregulated in both beta-cell lines and islets of mice when treated with high levels of stearic fatty acid (70,116).
Anti-apoptotic miRNAs fail in their homeostatic role when their expression is downregulated in glucolipotoxic environments. This is the case for miR-299-5p, whose expression is downregulated in vivo and in vitro in glucolipotoxicity environments. However, its overexpression is sufficient to protect beta-cells from high glucose/palmitate-induced cell death by downregulating the expression of its target genes, including some apoptosis-related genes Perp, Siah1, Rassf6, and Sgk1. Conversely, the selective inhibition of miR-299-5p results in a dramatic beta-cell dysfunction with impaired GSIS and significantly increased expression of apoptosis markers (71). miR-344-5p is another protective miRNA whose levels is downregulated in hypercholesterolaemia, and palmitate-induced lipotoxic diabetic beta-cells. Overexpression of miR-344-5p in beta-cells attenuates the lipotoxicity-induced beta-cell dysfunction and lipid accumulation and decreases cell apoptosis. Its inhibition aggravates the beta-cell dysfunction. miR-344-5p directly targets Cav1, initiating lipotoxicity-induced beta-cell death through caspase-3/Bax, MAPK/ERK and AKT signalling pathways (82).
Other interesting examples of anti-apoptotic miRNAs include miR-296-3p-whose expression is reduced in the serum of T2D, which protects against uric acid-induced apoptosis and beta-cell dysfunction by targeting Pten (69). miR-433, downregulated in the beta-cells treated with high glucose, also exerts a protective effect. miR-433 overexpression protects beta-cell viability from high glucose levels by inhibiting cell apoptosis and promoting cell proliferation. Mechanistically, miR-433 directly targets the 3'UTR of Cox2, suppressing its expression (93). miR-552-3p is also downregulated in the serum of T2D patients, exerting a homeostatic role by targeting and suppressing the expression of Jak1, and subsequently inhibiting the phosphorylation of STAT3, restraining the inflammation and apoptosis of beta-cells. Curiously, miR-552-3p is sponged by CircPIP5K1A. Thus, low levels of CircPIP5K1A decrease ROS, inflammation and apoptosis in cells exposed to glucolipotoxic stimuli, and the effect is reversed by the silencing of miR-552-3p (99).
Other miRNAs downregulated by hyperglycemia contributing to beta-cell failure include miR-532-5p, miR-383 and miR-409-3p. Specifically, miR-532-5p alleviates oxidative stress and inhibits cell apoptosis when cultured with high glucose, improving GSIS. These effects are mediated by directly targeting Ccnd1 and subsequent downregulation of p53 (98). miR-383 protects beta-cells from glucose-induced apoptosis and oxidative stress, and its overexpression in diabetic mice reverses HFD-induced hyperglycemia and pancreatic apoptosis. At the molecular level, miR-383 targets Tlr4, an nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway upstream activator, and ApoC3, a regulator of lipid metabolism (91). Another miRNA, miR-409-3p inhibits pancreatic beta-cell apoptosis by directly targeting the pro-apoptotic gene caspase-8. Interestingly, the Hsa_circ_0054633, whose expression is upregulated by high glucose, has been identified as a sponge for miR-409-3p (92).
Other miRNAs are upregulated in obesity and diabetes, but this does not imply their pathogenic contribution. For instance, miR-96 is a miRNA whose expression is upregulated in the islets of obese and diabetic patients or obese mice. However, upregulation seems to contribute to maintaining the system’s homeostasis since its overexpression in beta-cells promotes cell proliferation and inhibits apoptosis due to the negative direct regulation of beta-cell homeostasis regulators Foxo1 and Sox6 expression. In line with these findings, miR-96 KO mice show impaired beta-cell function in response to HFD (110).
There are also other miRNAs whose expressions are pathogenically relevant, causing a harmful phenotype promoting cell death, mediated or not by the regulation of BCL2 family members. This is the case for miR-101a, and miR-30b. Both are upregulated in beta-cells in response to interleukin (IL)-1b, promoting IL-1b-induced beta-cell dysfunction and apoptosis. These effects are mediated by downregulating the expression of the anti-apoptotic protein BCL2 by two different mechanisms: (I) miR-30b directly targets the 3'UTR in Bcl2 mRNA, and (II) miR-101a downregulates the expression of Bcl2 through the suppression of STAT3 (13).
miRNAs can promote beta-cell apoptosis through a pleiotropic repertoire of mechanisms. For instance, miR-375 is selectively expressed in human and mouse pancreatic islet cells and is upregulated in the pancreas and serum of T2D patients (117). Its direct targets are Psen1, myotrophin (V1) and Yap1 (45,84,86,87,89). Also, recent studies have shown that beta-cells export miR-375-3p into high-density lipoprotein (HDL) (88). Overexpression studies demonstrate that miR-375 induces apoptosis in beta-cells and inhibits cell proliferation. Conversely, loss of function protects beta-cells from glucolipotoxicity-induced cell death. The mechanism of miR-375 is diverse as it is positively correlated with islet amyloid and negatively correlated with islet mitochondria density (90). Moreover, the overexpression of miR-375 also reduces glucose-induced insulin gene expression and secretion by directly targeting Pdk1, decreasing the phosphorylation of PKB and GSK3α/β, downstream of PDK1 in the beta-cell insulin signalling pathway (83,85). miR-375 overexpression in vitro promotes beta-cell differentiation by targeting genes in several pathways, including some transcription factors such as Hnf1βb, Pax6, Insm1, Gata6, Pdpk1 and Insr (85). These are examples of miRNAs that exert control over the expression of genes responsible for different aspects of beta-cell function, synergising in their detrimental/beneficial effects on global beta-cell homeostasis.
miR-200 family miRNA coordinates the expression of multiple genes—this family consists of five members miR-141, miR-200c, miR-200a, miR-200b and miR-429. Curiously, the expression of miR-200 family members in mouse islets is not altered at prediabetic stages but significantly upregulated late in the natural history of the disease, when mice become severely diabetic. Interestingly, in human islets from T2D, only miR-200c is upregulated. The beta-cell-specific overexpression of miR-141/200c mice exhibits a diabetic phenotype with beta-cell dysfunction and apoptosis. Conversely, beta-cell-specific ablation of miR-141/200c protects from streptozotocin (STZ)-induced beta-cell death ameliorating T2D. At the molecular level, miR-200c directly binds and downregulates multiple genes such as the chaperone Dnajc3, the caspase inhibitor Xiap, Jazf1 (has a locus related to T2D), Rps6kb1(a downstream effector of mTOR and Bax inhibitor) and Ypel2, also positively controls tumour suppressor Trp53 activation, inducing the transcription of genes involved in pro-apoptosis pathways (49). Of note, inhibiting miR-200c biogenesis by the small molecule TGP-200c reverses its pro-apoptosis effect in beta-cells (118). Curiously, miR-200c has also been reported to regulate GSIS (see the section below). Another example of miRNA upregulated by diabetes is miR-335-5p, which inhibits pancreas beta-cell proliferation and promotes beta-cell apoptosis. miR-335-5p directly targets Slc2a4 (Glut-4) (77). Other researchers have also found that miR-335-5p suppresses pancreatic islet beta-cell secretion by directly inhibiting Vash1, and subsequently activating TGF-β signalling pathway in an in vivo model of gestational diabetes (119).
miR-30d is another miRNA that promotes beta-cell apoptosis while inhibiting proliferation and is downregulated in diabetic individuals and mice models. Its overexpression in beta-cells leads to hyperglycemia, reduced insulin level and glucose intolerance in mice fed HFD. Interestingly, miR-30d negatively affects beta-cells identity by downregulating beta-cell markers and inducing the expression of alpha-cell markers (73).
All in all, there is growing evidence of the promiscuity exerted by single miRNAs regulating the expression of multiple genes that control specific but complementary regulatory nodes or end effectors related to beta-cell physiology.
Another example of miRNA promoting beta-cell apoptosis is miR-146, which is upregulated in beta-cell lines treated with palmitate and the islet of db/db mice (30) and in T2D patients with poor blood glucose control (29). miR-146 overexpression promotes beta-cell apoptosis (30). miR-770-5p targets Triap1, a Tp53-regulated inhibitor of apoptosis, although this latter has only been found in association with gestational diabetes for the moment (120). miR-217 is upregulated in the serum of T2D patients and mouse islet beta-cells treated with high glucose. Overexpression of miR-217 in beta-cells inhibits cell proliferation and promotes high glucose-induced cell apoptosis, while depletion of miR-217 protects beta-cell from glucotoxicity and represses NF-κB signalling pathway. miR-217 directly targets Mafb (57). miR-708, whose expression is upregulated by endoplasmic reticulum (ER) stress and obesity, impairs beta-cell function and induces cell death in an effect mediated by the direct dysregulation of Nnat, an intracellular Ca2+ regulator on ER membrane (105).
Other interesting examples of detrimental miRNA are miR-338-3p and miR-27a, causing apoptosis, whose expression seems to be regulated at the molecular level by glucagon-like peptide 1 (GLP-1). The inhibition of miR-338-3p (whose expression is downregulated in genetically and dietary-induced obese mice) promotes beta-cell proliferation and protects beta-cell from apoptosis, while its overexpression has opposite effects, mimicking the effect of diabetes. Mechanistically, GLP-1 negatively controls the expression of miR-338-3p (79). Similar is the case for miR-27a, whose overexpression reverses the beneficial effects of GLP-1 on protecting beta-cells from cholesterol-induced cell apoptosis and lipid accumulation (67).
Lastly, some examples of miRNAs acting as CirRNA or lncRNAs’ intermediates promoting beta-cell apoptosis. Hence, miR-145-whose expression is decreased in the serum of T2D rodents-mediates the effect of CircANKRD36 inducing apoptosis. The downstream target of miR-145 is Xbp1, and silencing CircANKRD36 in diabetic mice results in the upregulation of the serum levels of miR-145 (27). miR-181a-5p also mediates the effects of the lncRNA PVT1 in INS-1 cells damaged by STZ (40).
miRNAs regulate beta-cell differentiation
The optimal differentiation from stem cells to beta-cells is critical for their mature functionality in maintaining glucose homeostasis. There is growing interest in cell therapy approaches transplanting stem cell differentiated beta-cells to restore pancreatic endocrine function in diabetes (121,122). Thus, understanding the early and late processes controlling beta-cell differentiation is an attractive research topic, including miRNAs’ contribution to homeostatic and pathological processes.
Specific miRNAs contribute to the early and late stages of differentiation (Figure 1). For instance, miR-7 is involved in the early stages of beta-like cell differentiation from human induced pluripotent stem cell (hiPSC) by targeting Gata6, Irs-2, Irs-1, Pax6, Pax4, NeuroD1, and Nkx2.2. The overexpression of miR-7 rapidly induces the expression of pancreatic transcription factors (insulin, Ngn3, Glut2, Pax4, Pax6, Kir6.2, Nkx6.1, Pdx1, glucagon, and Oct4) (101). Conversely, miR-181c-5p is more relevant at late hiPSC-derived beta-cell differentiation stages. The overexpression of miR-181c-5p increases PDX1 and NKX6.1 expression in pancreatic progenitors and increases the number of differentiated insulin-positive beta-like cells. miR-181c-5p directly binds to the 3'UTR of Smad7 and Tgif2 mRNA, repressing their expression and subsequently increasing the phosphorylation of SMAD2/3 and activating the TGF-β-SMAD2/3 signalling pathway. In line with these positive effects on beta-cell differentiation, mice transplanted with miR-181c-5p overexpressed hiPSC presents increased C-peptide secretion and improved glucose tolerance (41).
Other miRNAs reported to exert beneficial effects on beta-cell differentiation are miR-32 and miR-26. Overexpression of miR-32 promotes the expression of islet beta-liked cell markers and embryonic stem cell markers by directly downregulating the 3'UTR of Wwp2—an E3 ubiquitin ligase family member and subsequently inhibits ubiquitination and degradation of the embryonic stem cell transcription factor OCT4 (76). Concerning miR-26, it also promotes pancreatic endocrine cells differentiation, by targeting Tet1/2/3 and Tdg-and its overexpression in vivo results in increased postnatal islet numbers with higher expression of differentiation (Ngn3, Nkx6.1 and Pdx1) and endocrine markers (Ins1, Ins2, Gcg, Ppy, and Sst) (66).
miRNAs can also inhibit beta-cell differentiation. For instance, miR-21—whose expression is downregulated in insulin-producing cells (IPC) compared to pancreatic progenitor cells (PPC)—inhibits the differentiation from PPC into IPC and reduces insulin secretion by targeting Sox6—a negative IPC differentiation and function regulator, and Rbpj—a NOTCH signalling pathway downstream effector, and subsequently downregulating Rbpj’s target Hes1—a repressor of beta endocrine cell development (54). Another negative regulator is miR-145 which binds and silences the expression of lncRNA-ROR and Sox2. IncRNA-ROR competitively binds to miR-145, therefore maintaining the expression of Sox2. Interestingly, the reduction of IncRNA-ROR suppresses the pluripotency of stem cells, caused by the decreased expression of Sox2, Oct4, and Nanog and also impaired the differentiation and function of HuAEC-derived β islet-like cells, leading to beta-cell impaired functionality and lower glucose-induced C-peptide secretion (28).
miRNAs control insulin biosynthesis
miRNAs can also regulate the expression of the insulin gene. Specific miRNAs can alter (I) the transcription of the insulin gene to mRNA and/or (II) affect the transcription of genes relevant for the transcriptional regulation of the insulin mRNA or (III) the stabilisation of the mRNA transcript levels contributing to the fine-tuning of the insulin biosynthesis in healthy stages and also to the pathophysiological adaptations to disease.
One example is miR-30a-5p, whose expression is induced in the islets of db/db mice and beta-cells exposed to glucotoxicity conditions. In vitro overexpression of miR-30a-5p decreases the expression of the insulin gene, while its inhibition reverses the negative effect of glucotoxicity on insulin levels. Inhibiting miR-30a-5p in the pancreas of db/db mice increases insulin gene expression and insulin content, improving glucose tolerance. Mechanistically, miR-30a-5p directly targets the 3'UTR of the transcription factor NeuroD1 (72). Interestingly, another miR-30 family member, miR-30d, has the opposite effect of increasing glucose-induced insulin-gene transcription when overexpressed in vitro. The molecular mechanism for this opposite effect to miR-30a-5p is unknown, but NeuroD1 and Pdx1 seem to be discarded (74).
Another miRNA with a deleterious effect on beta-cell function is miR-802. This miRNA increases its expression in the islets of obese mouse models and impairs insulin transcription by targeting NeuroD1 and insulin secretion by targeting Fzd5, which is involved in calcium signalling (106). miR-19b, miR-101a, miR-30b and miR-124a are other miRNA known to negatively regulate the insulin gene transcription by directly interacting with the transcription factor NeuroD1 (13,14,39). Interestingly for miR-124, the transcription factor Ets2 has been shown to mediate the effects of epidermal growth factor (EGF) inhibiting miR-124a expression (15).
The cluster of miR-221/222 also exerts a negative effect on insulin biosynthesis. Their overexpression targets the 3'UTR of Nfatc3, causing impaired insulin production due to reduced insulin mRNA, protein content, and insulin vesicles (59). This contradicts other results in INS-1 cells showing that the overexpression of miRNA-221 stimulated insulin secretion and cell proliferation and suppressed apoptosis by directly targeting Pak1 (60). Similar discrepancies have been found for miR-204, which has been reported to suppress insulin biosynthesis by directly targeting Mafa (52). Nevertheless, Marzinotto et al. found no correlation between miR-204, Mafa and insulin expression. Moreover, the 3'UTR of Mafa is not predicted as a target of miR-204 (51). More research would be needed to clarify these contradictory results.
Other miRNAs directly increase insulin mRNA expression by altering the expression of signalling molecules and transcription factors (Figure 1). For instance, miR-483 targets 3'UTR of Socs3, repressing its expression and provoking an increase in insulin mRNA levels in beta-cells (96). In addition, miR-483 targets Aldh1a3, a marker of beta-cell differentiation protecting beta-cell function (97). Similarly, miR-30d increases the expression of the insulin transcription factor Mafa by interacting with Map4k4 3'UTR and inhibiting its expression, therefore inducing insulin expression and protecting beta-cells from inflammatory cytokine-induced dysfunction (75). The overexpression of miR-155-5p also increases insulin mRNA expression and improves glucose and insulin sensitivity. The mechanism of miR-155-5p involves in is binding the 3'UTR and inhibiting Mafb expression, which (I) promotes β-cell function; (II) induces the transcription of IL-6 in beta-cells; and (III) increases GLP-1 production in α-cells. Interestingly, miR-155-5p is upregulated in the islets of mice by hyperlipidemia-associated endotoxemia. The KO of miR-155-5p in hyperlipidemic mice causes beta-cell dysfunction (e.g., decrease in fasting insulin levels, decreased insulin content and decreased GLP-1 level) (36). Another exciting example is miR-205-5p, a miRNA whose expression is upregulated in the islets of the obese and diabetes-susceptible NZO mice (a polygenic model of obesity and diabetes). The overexpression of miR-205-5p increases insulin 1 mRNA expression and intracellular insulin content. Mechanistically, miR-205-5p directly interacts with the Tcf7l2 gene, a transcription factor that regulates genes involved in Wnt signalling and beta-cell function. Other predicted targets for miR-205-5p include Plcb1, Nphp1 and Cxxc4, and Plcb1, whose expression is significantly downregulated in beta-cells when miR-205-5p is overexpressed (53).
miRNAs directly binding to the insulin gene have also been described. One is miR-196b, which positively regulates the biosynthesis of insulin 2 by directly targeting the 5'UTR of insulin 2 mRNA and activating the translation of insulin 2 in the presence of Ago2. It is worth mentioning that miR-196b can displace the RNA binding protein HuD by competing for the same binding site, thus abolishing the inhibitory impact of HuD on insulin 2 translation (47). This reinforces previous evidence that miRNAs can compete or synergise with other RNA regulators and miRNA over the same targets.
The opposite effect has been described for miR-25 and miR-92a. The overexpression of these two miRNAs decreases the expression of proinsulin, insulin content and secreted insulin, superseding the inhibitory effect of miR-9 in insulin secretion. Both bind directly to 3'UTR of Ins1 mRNA, overlapping with the one for Ptbp1, a protein that stabilises insulin mRNA (65). Another interesting example is the miR-133a, whose expression is upregulated in the blood of prediabetic and T2D patients (22). miR-133a also directly targets Ptbp. Remarkably, human islet cells treated with the miR-133a precursor presents reduced insulin levels (23).
miRNAs control insulin secretion
The last process involving insulin production is insulin protein being transported in vesicles to the plasma membrane and released. A significant number of miRNAs (Figure 1) regulate insulin exocytosis by modulating the expression of genes responsible for the transport, the process of docking and the fusion of vesicles to the plasma membrane, or intermediate hubs that control the expression of these genes (123).
miR-212 and miR-132 positively regulate GSIS under the regulation of GLP1 (20). Increased intracellular cAMP elevates the expression of miR-212 and miR-132 in a cAMP-SIKs-CRCT1 dependent manner. Crct1 is predicted to be the direct target of these two miRNAs, as part of negative feedback in beta-cells to maintain intracellular homeostasis. In vivo beta-cell-specific overexpression of miR-212 or miR-132 improves insulin secretion and glucose homeostasis and enhances the proliferation of beta-cells in the mice when fed HFD (miR-312 only) (21). Of relevance, their expression is upregulated in the islets of obese, prediabetic and diabetic mice models and in beta-cells cultured with high glucose and palmitate, which can be interpreted as part of the regulatory system ensuring appropriate levels of insulin to counteract the insulin resistance stage (56,124).
Another miRNA recently reported to stimulate GSIS is miR-21. This miRNA is upregulated in islets of diabetic db/db mice and mice fed with high fat and high glucose diets. Notably, it promotes GSIS by upregulating the expression of Glut2 via a miR-21-Pdcd4-AP-1 axis. Thus, miR-21 specifically delivered into the pancreas of diabetic db/db mice seems to improve blood glucose and insulin secretion, and conversely, the beta-cell-specific KO mice for miR-21 shows glucose intolerance and decreases GSIS (55).
Overexpression of miR-139-5p in beta-cells decreases beta-cell function. miR-139-5p acts as an upstream negative regulator of PICK1, a membrane protein involved in insulin granules formation and maturation (125), interacting with its 3'UTR. Overexpression of Pick1 could reverse the adverse effect of miR-139-5p on beta-cell, as Pick1 contributes to the activation of the PI3K/Akt pathway and increases the expression of Glut2 and also promotes insulin secretion (24).
Opposite to the previous miRNAs, miR-124a expression is elevated in T2DM patients’ islets, and its overexpression impairs GSIS. Confirmed targets for miR-124a are Mtpn and Foxa2, two insulin secretion regulators (14). Similarly, miR-187 is upregulated in islets from T2D patients, and its expression negatively correlates with GSIS in non-diabetic human islets. Hipk3, an insulin secretion regulator, is identified as its direct target, and the overexpression of miR-187 has been shown to impair GSIS in rat primary islets and beta-cell lines (44).
miR-153 expression is also increased in islets of obese and diabetic mice, and its overexpression inhibits both basal and GSIS as a result of a significant decrease of docked insulin granules in vitro, resulting in impaired glucose tolerance in vivo. miR-153 directly targets the mRNAs of some SNARE proteins (including Vamp2, Snap25 and Stx1a). Thus, the inhibition miR-153 or the overexpression of SNAREs family members could recover the insulin secretion in a diabetic setting (34). Similarly, miR-29a, whose expression is upregulated in beta-cells in response to high glucose concentrations, inhibits insulin exocytosis by decreasing Stx1a (a two-SNEARs protein) mRNA by directly targeting its 3'-UTR (68). Also, miR-34a expression is upregulated in beta-cell lines treated with palmitate, in the islet of db/db mice (30) and overweight/obese T2D patients (29). miR-34a overexpression impairs GSIS and induces beta-cell apoptosis by targeting the SNARE family member Vamp2 and the anti-apoptotic factor Bcl2. Increased miR-34a is also linked to the activation of p53, which could contribute to apoptosis. miR-34a also targets Sirt1 and exacerbates ROS accumulation and apoptosis. Conversely, the inhibition of miR-34a reduces apoptosis induced by high glucose and palmitate (81).
Another inhibitor of insulin secretion is miR-9, which is selectively expressed in islets and the brain. The overexpression of miR-9 in beta-cells decreases insulin exocytosis. The direct targets of miR-9 include three genes that span different roles related to insulin secretion Sirt1, Oc2 and Stxbp1. Thus, Sirt1 promotes insulin secretion by inhibiting UCP2 expression and Oc2 that directly binds to the promoter of granuphilin, a Rab3/Rab27 effector that inhibits insulin secretion. Finally, Stxbp1 plays a fundamental role in the fusion of the secretory granules containing insulin with the plasma membrane (107,108).
Interestingly, miR-335 is another miRNA negatively associated with insulin secretion in human islets from prediabetic individuals, and its expression is upregulated in the GK rat, a non-obese T2D model rodent model (126). The overexpression of miR-335 results in decreased GSIS due to decreased rapid exocytosis, despite having no effects on the docking or altering Ca2+ current. The molecular targets of miR-335 are three exocytotic proteins: SNAP25, STXBP1 and SYT11, whose expressions are downregulated by miR-335 (78).
miR-218 and miR-322 also target Stxbp1 (as for miR-9 and miR-335). Their expression is downregulated after a prolonged glucose treatment, inhibiting insulin release (58). Another exciting example is miR-200c, previously discussed to regulate apoptosis. miR-200c targets Etv5, a transcription factor that regulates the expression of insulin exocytosis genes, including Syt11, Aqp3 and Cbfb. The knockout of miR-200c in islets from T2D patients significantly improves GSIS (50).
miR-7a is another miRNA highly expressed in human and mouse islet cells whose expression is initially downregulated in the islets of genetic and diet-induced obese mice at early stages, although its expression seems to increase in advanced stages (102). Overexpression of miR-7 in islets beta-cells of mice leads to a diabetic phenotype by directly targeting the 3'UTR of Myrip mRNA and subsequently inhibiting insulin granule transportation and release (104) and by targeting Reg1, a gene associated with the prevention of apoptosis (127). miR-7a2, a dominant form of miR-7 in the pancreas, controls the late stages of insulin granule fusion with the plasma membrane and ternary SNARE complex activity. At the molecular level, the direct targets of miR-7a2 include vesicle fusion and SNARE activity (Snca, Cspa, Cplx1), calcium-regulated vesicular trafficking (Pkcb), cytoskeleton rearrangement (Pfn2, Wipf2, Basp1, Phactr1), and membrane targeting (Zdhhc9). Briefly, miR-7a2 binds 3'UTR of Snca, reducing its expression, disrupting the interaction between SNCA and VAMP complex, and decreasing SNARE activity (102).
miR-153, a miRNA located in the gene Ia-2β, suppresses glucose-/potassium-induced insulin secretion in beta-cell line and primary mice islet cells by directly targeting the 3'UTR of the RNA that codes for the calcium channel gene Cacna1c, which is physically coupled to the genes related to vesicle release including RIMs, Snap-25 and Syntaxin-1. Interrupting the interaction between miR-153 and Cacna1c abolishes the effect of miR-153 in GSIS. Knockdown of the hosting gene Ia-2 or/and Ia-2β mice beta-cells shows fewer dense-core vesicles (DCV) by decreasing their half-life and reducing insulin secretion (35).
miR-145 overexpression decreases GSIS and cholesterol efflux in beta-cells. miR-145 directly binds the 3'UTR of the cholesterol transporter Abca1 and suppresses its expression. Other miRNAs such as miR-148, miR-27, miR-144, and miR-33a/33b also target and suppress the 3'UTR of Abca1 mRNA. Interestingly, the specific Inhibition of Abca1 in the pancreas causes impaired insulin secretion and higher insulin content in beta-cells, suggesting that the disturbances in cholesterol are the main effector of such disfunction (26,128). Interestingly the non-coding RNA lnc-DANCR has been shown to act as a sponge for miR-33a, a miRNA involved in gestational diabetes mellitus by inhibiting insulin production (26). miR-463-3p is upregulated in the islet of T2D patients and negatively correlates with GSIS. The direct target of miR-463-3p is Abcg4 (95). miR-463-3p overexpression in beta-cell line and in vivo pancreas inhibits GSIS, while its inhibition promotes insulin secretion.
miR-24 expression is upregulated in response to oxidative stress and/or cholesterol accumulation, and it is present in high levels in islets from db/db and HFD-fed mice. Interestingly, when overexpressed in islets, it impairs GSIS. Mechanistically, miR-24 reduces the expression of Sp1, causing the inhibition of transcription of Scgn. Reduced levels of Scgn downregulate the phosphorylation of the focal adhesion kinase and paxillin, reducing the exocytosis of insulin granules. Also, miR-24 directly targets transcription factors Hnf1α and Neurod1, contributing to the impairment of GSIS and defects in beta-cell proliferation (64). Intriguingly, in the presence of high cholesterol, the inhibition of miR-24 rescues the insulin1 miRNA expression, but its mechanism remains unclear (63).
These findings evidence a crosstalk between the levels of cholesterol and miRNAs contributing to the impairment in insulin biosynthesis/secretion, raising the possibility that defective lipid metabolism, lipotoxicity—a hallmark of obesity and associated comorbidities such as diabetes and fatty liver—might do so through triggering the inappropriate expression of miRNAs.
Other miRNAs regulating insulin biosynthesis and secretion include miR-577, whose expression is upregulated in the blood of diabetic children. Interestingly, the overexpression of miR-577 in embryonic beta-cells blunts the effects of Fgf21 inducing insulin biosynthesis and secretion by directly targeting Fgf21 and inhibiting Fgf21 mediated activation of ERK1/2 signal (100). miR-125b is another miRNA upregulated in mouse and human islets in response to high glucose, and its overexpression in vivo and in vitro has impaired insulin production and secretion from beta-cells. Interestingly, the targets of miR-125b include the genes encoding lysosomal protein M6PR and mitochondria homeostasis regulator MTFP1 (16), evidencing the role of this miRNA regulating organelle dynamics that contribute to beta cell function.
Let7b-5p also exert this dual effect, but in this case, biosynthesis and secretion are regulated in opposite directions. Let7b-5p is highly expressed in the serum of T2D patients, and its overexpression increases insulin biosynthesis but paradoxically inhibits GSIS, while let7b-5p inhibition decreases insulin content and partially restores GSIS. On a molecular level, let7b-5p plays a role in dysregulating the transcriptional level of Insulin, Pdx1, Gck, Glut2 and Insr (111).
A very intriguing mechanism through which several miRNAs seem to control insulin secretion consists of regulating the intracellular bioenergetics. Specifically, miR-15a and miR-130a/miR-130b/miR-152 target intracellular adenosine triphosphate (ATP) production, causing impairment of ATP-dependent insulin production and secretion. Moreover, miR-15a downregulates Ucp2 expression by targeting its 3'UTR directly, causing changes in mitochondrial performance, uncoupling ATP production from oxygen consumption in pancreatic beta-cells, and promoting insulin biosynthesis (32). miR-130a/miR-130b/miR-152 act differently, decreasing the ATP production by indirect (miR-130a/miR-130b)/direct (miR-152) interaction with ATP production protein Pdha1. These three miRNAs can also reduce Gck expression and negatively affect ATP-dependent insulin production and secretion (19).
miRNAs in type 1 diabetes
T1D is a chronic autoimmune disease characterised by infiltrated immune cells, eventually leading to beta-cell death (129). A priori, there are two ways how immune cells cause adverse effects on beta-cells: (I) the pro-inflammatory mediators such as cytokines released from macrophages and T lymphocytes that induce ER stress and beta-cells apoptosis (130); (II) the release of exosomes produced by immune cells that target beta-cells (131). Recently, a growing body of evidence highlights the critical roles of miRNAs in regulating the autoimmune response in T1D, aiming to provide potential therapeutical targets to protect beta-cells against immune cell attacks.
Several miRNAs regulate immune cell activation in the context of T1D. Thus, for example, the expression of miR-20a and miR-326 are upregulated in peripheral blood mononuclear cells (PBMCs) of T1D patients. Interestingly, the predicted targets of miR-20a and miR-326 are genes classically associated with T1D and other autoimmune diseases (132). For example, miR-326 targets Ets1 and is predicted to target Vdr, both genes involved in modulating the immune response (133).
miR-202-3p is upregulated in pancreas-infiltrating lymphocytes (PILs) compared to other immune cells such as thymocytes and peripheral T CD3+ lymphocytes. Interestingly, miR-202-3p targets Ccr7 or Cd247 mRNA in the PILs of non-obese diabetic (NOD)-mice, contributing to the regulation of autoimmune response in T1D (134). Similarly, miR-142-3p is upregulated in peripheral blood T cells from T1D human patients and NOD mice, an animal model for studying autoimmune diabetes. Thus, the miR-142-3p aggravates high glucose-induced inhibition of Treg cells induction, whereas a miR-142-3p inhibitor promotes Treg induction. Thus, the inhibition of miR-142-3p seems to attenuate the islet autoimmunity in NOD mice. The miR142-3p/Tet2/Foxp3 axis is responsible for enhancing Treg induction and stability. miR-142-3p directly targets and negatively regulates Tet2, a regulator involved in Treg induction and maintenance, by activating demethylation of the Foxp3 CNS2 (135). miR-216a is another example of miRNA whose expression is induced in NOD mice. Interestingly, miR-216 promotes pancreatic beta-cell proliferation by directly targeting and suppressing the Pten (136). miR-21 is another miRNA upregulated in islets of the NOD mice and inflammatory cytokine-treated beta-cells. miR-21 is transcriptionally activated by NF-κB family members c-Rel and p65 and prevents beta-cell apoptosis in the context of T1D by negatively controlling the expression of Pdcd4, an apoptosis inducer via Bax family (137). miR-203a is another example of miRNA upregulated in pancreatic beta-cells of NOD mice. Elevated miR-203a inhibits proliferation and promotes apoptosis in MING6 cells by targeting Irs-2. Conversely, a miR-203a inhibitor ameliorates the hyperglycemic phenotype of NOD mice (138).
The miR-17-92 family members promote lymphomagenesis disorders and autoimmunity (139). One of the family members, miR-92a, is highly expressed in (CXCR5)+ CD4+ T follicular helper (TFH) cells at the early onset of islet autoimmunity, and its levels correlate with insulin autoantibody (IAA). In vitro, miR-92a induces TFH precursor induction, while inhibition of miR-92a suppresses TFH precursor induction but increases Treg cells induction. Inhibition of miR-92a in the context of NOD mice shows decreased TFH precursors in peripheral blood or islet lymph nodes, limiting the activity of islet autoimmunity. The mechanism of miR-92a includes (I) Klf2 as a direct target of miR-92a; (II) miR-92a inducing TFH precursor is abolished by PI3K inhibitor but is aggravated by PTEN inhibitor, evidencing the role of PTEN-PI3K signalling pathway in mediating miR-92a effects (140).
Interestingly, many miRNAs have been reported as cargo of exosomes released by immune cells acting on beta-cells in T1D. For instance, in NOD mice, miR-142-3p, miR-142-5p and miR-155 are expressed and released from T lymphocytes and transferred to islet beta-cells via exosomes. Mechanistically, these miRNAs trigger beta-cell apoptosis and increase the expression of genes involved in chemokine signalling, including Ccl2, Ccl7, and Cxcl10. In line with this evidence, exosomes produced by cultured T cells from NOD mice increase the translocation of NF-κB and promote apoptosis in beta-cells (141). Conversely, inhibiting these miRNAs in vivo reduces beta-cell dysfunction and inflammation and improves diabetes.
It is worth noting that several miRNAs are downregulated in both T1D patients and preclinical models of T1D. For instance, miR-16-5p expression is downregulated in the plasma of T1D patients. The overexpression of miR-16-5p in beta-cells increases cell proliferation and reduces apoptosis in response to high glucose. Mechanistically, miR-16-5p negatively regulates Cxcl10, and conversely, the treatment with CXCL10 reverts the beneficial effects of miR-16-5p overexpression. The overexpression of miR-16-5p also increases the expression of Bcl2 and decreases Bax and cleaved Caspase-3 levels (142). miR-150 is also downregulated in T1D patients’ islet beta-cells. Moreover, NF-κB activates miR-150 via direct interaction between NF-κB and miR-150 promoters. Inhibition of miR-150 could reverse the effects of NF-κB, promoting beta-cell apoptosis and inflammation. miR-150 targets and suppresses Puma expression, inhibiting inflammation and beta-cell apoptosis induced by T1D (143). miR-125b-5p is downregulated in an experimentally induced diabetes model using HFD + STZ. Interestingly, its overexpression promotes beta-cell proliferation and inhibits cell apoptosis. miR-125b-5p acts on Dact1 and inhibits the JNK signalling pathway (144). Another example is miR-409-3p, a miRNA enriched in CD8+ central memory T cells whose expression gradually declines in the blood and pancreas of T1D patients and T1D mouse models, which positively correlates with the severity of insulitis (145).
Of particular interest are miRNAs reported to play a role in both T1D and T2D pathogenesis, contributing to shared processes at the molecular level. One key example is the miR-29 and miR-26 family members. In T1D, miR-29a/b/c are upregulated in the beta-cells (not in infiltrated immune cells) of the islets of NOD mice and human and mouse islet beta-cells treated with pro-inflammatory cytokines. In T2D, miR-29 family members are upregulated in obese and diabetic mice, and glucose levels induce their expression in humans and rodents. The overexpression of miR-29 in beta-cells induces inflammation-induced apoptosis by directly targeting and reducing the expression of the Bcl2 family member Mcl1. Moreover, in vitro, miR-29 impairs GSIS by targeting the transcription factor Onecut2, which causes an increase of the insulin exocytosis inhibitor granuphilin (146,147). Paradoxically, the miR-29a KO exhibits a phenotype of T1D during unfolded protein stress and decreased insulin exocytosis (148). This apparent contradiction between in vivo and in vitro evidence the complexity of investigating the mechanisms associated with altered levels of miRNAs in health and disease. Of note, miR-29 is also enriched in the liver and is upregulated in the livers of ob/ob mice (149). The miR-29 family acts as a negative regulator of hepatic insulin sensitivity, and the global or hepatic miR-29a or miR-29c knockout are protected from diet-induced obesity and insulin resistance (148) by increasing the insulin signalling pathway via directly regulating PI3K expression (149).
Another T1D mouse model with an increased level of miR-26a in islets and T lymphocytes shows a prolonged normoglycemic period before T1D ensues, exhibiting lower grades of insulitis mediated by the inhibition of autoreactive CD4+ and CD8+ T cells and the expansion of Treg (150). Of relevance, miR-26a is downregulated in the islets and serum of obese and T2D diabetic mice, and its overexpression alleviates the phenotype of T2D by improving beta-cell function and peripheral insulin sensitivity. Thus, the overexpression of miR-26a protects mice from hyperinsulinemia induced by HFD, maintaining adequate insulin secretion and signalling. Mechanistically, miR-26a directly targets genes involved in beta-cell proliferation and insulin secretion, such as Pten, Cacna1c, Crebrf, Ctgf, Esr1, Ext1, Mtpn, Onecut2, Pfkfb2, Pja2, Plcb1, Rhoq and Sox5 (151,152). Moreover, miR-26a also enhances insulin sensitivity in peripheral organs transported in exosomes (see in section “miRNAs mediate cell to cell and organ to organ crosstalk”).
miRNAs in diabetes linked to pancreatitis
Post-pancreatitis diabetes mellitus (PPDM) is a frequent complication of pancreatitis, as hyperinsulinemia during pancreatitis might be responsible for the development of insulin resistance (IR) (153). Acute pancreatitis is an acute abdominal inflammatory disease associated with necrosis of the pancreas, systemic inflammation, multiple organ dysfunction, and mechanistically related to a cytosolic calcium overload (154) whereas chronic pancreatitis is a progressive syndrome characterised by pancreatic inflammation, leading to extensive tissue fibrosis and deficiency of pancreatic exocrine and endocrine function (155). Recent investigations have shown that in pancreatic acinar cells, miR-26a targets Trpc3 and Trpc6 store-operated Ca2+ entry (SOCE) channels and attenuates physiological oscillations and pathological elevations of intracellular calcium alleviating experimental pancreatitis (156). On a different note, miR-153 has been described to aggravate acute pancreatitis in hypertriglyceridemia by targeting Traf3 and delaying pancreatic repair. Interestingly, SREBP1c transcriptionally suppresses miR-153 and facilitates tissue regeneration in vivo (157), which opens a potential therapeutical window for these patients. In a similar line, chronic pancreatitis relates to chronic inflammatory and fibrotic phenotype with reduced beta-cell mass, where pancreatic stellate cells (PSC) are critical in the progression of fibrosis. In this case, several miRNAs mediate TGF-β1 effects in PSC. For instance, miR-34b mediates the induction of extracellular matrix accumulation (158), whereas miR-140-3p and miR-143-3p directly target Bcl2 to increase beta-cell apoptosis (159).
miRNAs mediate cell to cell and organ to organ crosstalk
Over the last decades, a growing body of evidence shows that extracellular vesicles (EVs)-/exosome-derived miRNAs mediate cell-to-cell crosstalk and organ-to-organ communication. This is also the case for the pancreas (160,161). However, the number of investigations focused on miRNAs’ para/endocrine role is still limited. Also, there is evidence that the endocrine pancreas can release exosomes containing miRNAs into the bloodstream and target specific tissues modifying their metabolic activities and vice-versa. Other metabolic tissues and organs also secrete miRNAs that target the endocrine pancreas to regulate its viability and/or functionality (Figure 2).
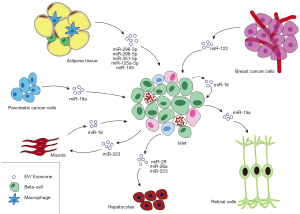
miRNAs in beta-cell to beta-cell crosstalk
Pancreatic beta cell-derived exosomes have been reported to contain +200 miRNAs, among them miR-7, miR-29a, miR-146a, miR-451 and miR-142-3p are known to be secreted from beta-cells in response to pro-inflammatory cytokines and transferred to neighbouring beta-cells affecting basic processes such as apoptosis, proliferation and the activity of recipient beta-cells (162).
miRNAs connecting beta-cells to hepatocytes
miR-29 is one of the first miRNAs described to mediate the crosstalk between beta-cells and macrophages. Briefly, miR-29 released by beta-cells promotes inflammation by recruiting and activating circulating macrophages (163). miR-29 increases in the serum of obese subjects and mice fed HFD under fasting conditions and directly in response to free fatty acid (FFA) treatment in vitro. Briefly, exosomes enriched in miR-29 originated in beta-cells impair insulin sensitivity in isolated hepatocytes by downregulating AKT phosphorylation and increasing glucose output (164). Similarly, the overexpression of exosomal miR-29 in vivo attenuates insulin sensitivity and increases blood glucose levels. Of relevance, the beta-cell-specific depletion of miR-29, by using a miRNA-sponge, reverses the HFD-induced insulin resistance phenotype in vivo.
Conversely, other miRNAs originating in the pancreas can play a beneficial role in liver metabolism. One of them is miR-26a, whose expression is downregulated in the serum of obese and T2D patients, in the islets of diet-induced or genetically obese mice, and in the liver of overweight humans. Interestingly, beta-cell-specific overexpression of miR-26a improves hepatic insulin sensitivity and glucose tolerance and decreases hepatic glucose production and fatty acid synthesis. Mechanistically, miR-26a targets the 3'UTR of critical regulators involved in liver fatty acid synthesis (Acsl3, Acsl4), gluconeogenesis (Pck1, Tcf7l2) and insulin signalling (mediating phosphorylation of IRS proteins Gsk3b, Pkcd, Pkcq) (152,165). Another exciting example is miR-223 which originates in beta-cells and whose expression is upregulated by high glucose. This miRNA has been reported to travel to the liver, and muscle in a mechanism mediated by exosomes and regulate the insulin signalling pathway of targeted organs by inducing the expression of Glut4 (166).
Globally considered, these findings provide robust evidence of miRNA-mediated crosstalk between the endocrine pancreas and liver and reinforce the notion that failure in the pancreas could exacerbate the phenotype of other organs in the context of metabolic diseases.
miRNAs in muscle and beta-cell crosstalk
EVs secreted by muscles contain numerous miRNAs, including miR-16, whose expression is upregulated in response to high palmitate diet-induced insulin resistance. Interestingly, this miRNA can be taken up by the pancreas, thus promoting beta-cells proliferation. At the mechanistic level, miR-16 targets and inhibits Ptch1, an Hh receptor in Hedgehog signalling pathway involved in pancreas development (167,168).
miRNAs in Beta-cells crosstalk to retinal cells
It has been reported that miRNAs secreted from the pancreas can contribute to diabetic retinopathy, a common complication resulting from the late stages of diabetes. This is the case for miR-15a, whose expression is upregulated in diabetic patients and mice plasma. Thus, exosomal miR-15a secreted from beta-cells is increased in response to high glucose stimulation and transported to the circulatory system regulating retinal function. Overexpression of miR-15a in Muller cells either by being transfected with miR-15a precursor or culturing them with the exosomes released by high glucose-treated INS-1 cells results in oxidative stress, mediated by direct interaction between miR-15a and Akt3, a downstream factor of PI3K in insulin signalling pathway (169), and ultimately leading to cell apoptosis (170).
miRNAs in Adipose tissue crosstalk to beta-cells
Healthy adipocytes secrete EVs (Ad-EVs) that can be taken up by beta-cells promoting beta-cells survival and repressing cell death. In contrast, EVs produced by adipocytes treated with proinflammatory cytokines (CK-EVs) have opposite effects on recipient beta-cell viability by regulating the critical signalling pathways, including Akt-GSK-3β, ERK-eIF2α, and c-JUN pathways (171), which are involved in cell proliferation, survival and function (172). Interestingly, the miRNA cargos in Ad-EVs and CK-EVs are different and modulate the inflammatory fingerprint in beta-cells (171).
miRNAs in immune cells crosstalk to beta-cells
M1 macrophages that accumulate in the islet of diet-induced obese mice, secrete exosome enriched with miR-212-5p that can be taken up by beta-cells, inhibiting beta-cell GSIS. Interestingly, the transfer of M1 macrophage-derived exosomes (M1-exos) to the pancreas of living mice causes impaired glucose tolerance and insulin secretion. At the molecular level, miR-212-5p targets Sirt2 3'UTR, inhibiting its expression and impairing SIRT2-mediated AKT/GSK-3β/β-catenin signalling pathway. Blocking miR-212-5p in M1-exos in vivo improves glucose tolerance in HFD-induced obese mice by alleviating the impaired beta-cell GSIS (173). Interestingly, adipose tissue macrophages (ATM) have also been reported to release EVs that regulate pancreatic beta-cell adaptation and dysfunction in the context of obesity. Thus, miR-155, highly enriched in obese ATM EVs, has been shown to act on Mafb in beta-cells (36,174).
Another example of how miRNA secreted from immune cells affects beta-cell homeostasis comes from studies conducted by Guay et al. where exosomes derived from T lymphocytes containing miR-142-3p, miR-142-5p, and miR-155 can be taken by islet beta-cells, inducing beta-cell apoptosis. Mechanistically, this occurs by the upregulation of genes involved in chemokine signalling (including Ccl2, Ccl7, and Cxcl10). The inactivation of those miRNAs using a “miRNA sponge” prevents beta-cells from exosome-induced cell death, thus preventing T1D development in pre-diabetic NOD mice (141).
On the other hand, bone marrow transplantation (BMT) promotes beta cell regeneration in STZ-induced diabetic mice in a mechanism mediated by bone marrow (BM) cells derived exosomes. Briefly, miR-106b-5p and miR-222-3p are secreted by BM cells and increased in pancreatic islet cells after transplantation. At the mechanistic level, these miRNAs suppress the expression of Cip/Kip family members p21Cip1 and p27Kip1, promoting post-injury beta-cells regeneration and improving glucose metabolism (175).
miRNAs mediate pancreatic cancer cells and beta-cells crosstalk
There is growing evidence showing how miRNA transported in exosomes derived from pancreatic cancer cells, or pancreatic cell lines can be transported and taken by beta-cells impairing GSIS. A relevant example is miR-19a which is enriched in pancreatic cells-derived exosomes. miR-19a directly targets 3'-UTR of Adcy1 [a critical enzyme in insulin secretion by producing cAMP (176)] and Epac2 [also known as cAMP-GEFII, mediates cAMP-dependent insulin secretion (177)] reducing cAMP and Ca2+ in recipient beta-cells (178). Of note, miR-19a also impairs beta-cell insulin secretion by directly targeting and inhibiting the expression of insulin gene transcription factor Neurod1 (179). As a proof of concept, mice transplanted with pancreatic cancer cells where miR-19a has been knocked-down exhibit improved glucose tolerance and insulin sensitivity (178). This evidence is critical for understanding pancreatic cancer-associated diabetes and paving new avenues for the design of therapeutical strategies.
miRNAs in Breast cancer cells crosstalk to islet beta-cells
miR-122 in EVs correlates with high fasting blood glucose and low insulin levels in breast cancer patients. Interestingly, miR-122 is produced by breast cancer cells and taken by pancreatic islet beta-cells, negatively regulating beta-cell ATP-dependent insulin exocytosis by directly targeting pyruvate kinase (Pkm) (180).
Conclusions
The development of new and more precise high-throughput analytical tools in the field of bulk and single-cell transcriptomics along with exosome biology and the availability of integrative bioinformatics tools is expanding our understanding of the causative role of miRNAs, providing a more detailed map of the miRNA mediated crosstalk between cells and organs in health and disease.
In this review, we summarised relevant research studies from the last eighteen years that integrate our current knowledge of how miRNAs affect islet function in the context of diabetes. We also show the emerging evidences of the crosstalk mediated by exosomes between pancreas and other peripheral organs and its potential contribution to the complex pathophysiology of diabetes.
We believe that using genetically modified models to selectively increase or decrease pools of miRNAs in a particular time/spatial frame (conditional tools) and a specific cell type will help dissect the relevance of miRNAs on global organismal homeostasis and metabolism. These experimental models, combined with the development of more accurate prediction algorithms and machine learning-based analysis, will assist in understanding the abnormal miRNA profiles observed in multiple metabolic diseases such as obesity and diabetes (181-183). Nevertheless, despite the extreme value of animal models, a cautionary note is needed as there is a growing body of evidence showing significant species-specific differences in the development (184), microarchitecture and transcriptomics of the pancreatic islets (185-188) and functional differences such as susceptibility to injury (189) that could limit the value of animal models on reflecting certain aspects of human islets pathophysiology. Novel examples of some of these interspecies limitations have been recently highlighted by two excellent reviews reporting the species-specific evidence for de-differentiation/altered activity of islet beta cells as mechanisms of failure and recovery during maturation and T2D (190,191) or the work conducted by Keller and Perez, where they identified human CREM as a target gene of miR-375, where the predicted binding site was conserved in humans and other primates, but not in species like mice or rats (192).
Another critical area of research will be finding the molecular mechanisms responsible for the transcriptional regulation of miRNA in the context of disease. Understanding how, where, why and when miRNAs act is an unmet need to evaluate the value of miRNA as prognostic or diagnostic tools in clinical practice. Few examples of this are for instance the recent integration of cis-eQTLs and GWAS data as a promising approach to identifying signals associated with T2D; in the latter, five miRNAs, miR-7-5p, miR-126-3p, miR-1236, miR-130b-5p miR-1275 and miR-194-5p have been recently identified as potential signals for T2D and/or insulin secretion rates (11), the discovery of SNPs in miR-196a and miR-423 linked to increased risk of T2D in Middle East regions (193) or the identification of “unique” signatures in the miRNA profile during the progression of T2D, where for instance, miR-222-3p and miR-148-3p were specifically modulated in diabetic patients and correlated with clinical parameters of glucose and lipid metabolism (194). Interestingly, miR-148-3p, also found to be increased in the serum of T1D (195), is highly present in human and cow milk exosomes and it has been hypothetised as a modulator of beta cell de-differentiation—at the transition that takes place during weaning process—and to play a pathogenic role in T2D during adulthood (196) adding a new perspective on the roles of dietary miRNAs as bioactive components of food and their relevance on health and disease (197).
Globally considered, we are confident that elucidating the regulatory networks controlling miRNAs and their targets will facilitate finding specific genes/signalling hubs of therapeutic significance. There are still significant limitations (e.g., promiscuity of targets, stability and target delivery) (198), and the limited number of clinical trials using miRNA. However, advances in engineering and refining miRNA therapeutics in metabolic diseases look promising and deserve further attention.
Acknowledgments
We thank Mr. Mark Campbell (Wellcome Trust-MRC Institute of Metabolic Science, University of Cambridge, United Kingdom) for his helpful suggestions during the preparation of this manuscript.
Funding: We thank the MRC MDU (MC_UU_12012/2) and Cambridge University Nanjing Centre of Technology and Innovation’s research grant for funding this work.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Chenyu Zhang) for the series “Extracellular RNAs and Human Health” published in ExRNA. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-17/rc
Peer Review File: Available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-17/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://exrna.amegroups.com/article/view/10.21037/exrna-22-17/coif). The series “Extracellular RNAs and Human Health” was commissioned by the editorial office without any funding or sponsorship. This work was funded by the MRC MDU (MC_UU_12012/2) and Cambridge University Nanjing Centre of Technology and Innovation’s research grant. AVP serves as an unpaid editorial board member of ExRNA. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Galicia-Garcia U, Benito-Vicente A, Jebari S, et al. Pathophysiology of Type 2 Diabetes Mellitus. Int J Mol Sci 2020;21:6275. [Crossref] [PubMed]
- Skyler JS. Diabetes mellitus: pathogenesis and treatment strategies. J Med Chem 2004;47:4113-7. [Crossref] [PubMed]
- Cerf ME. Beta cell dynamics: beta cell replenishment, beta cell compensation and diabetes. Endocrine 2013;44:303-11. [Crossref] [PubMed]
- Hudish LI, Reusch JE, Sussel L. β Cell dysfunction during progression of metabolic syndrome to type 2 diabetes. J Clin Invest 2019;129:4001-8. [Crossref] [PubMed]
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019;20:21-37. [Crossref] [PubMed]
- Ji C, Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol 2019;15:731-43. [Crossref] [PubMed]
- Isaac R, Reis FCG, Ying W, et al. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab 2021;33:1744-62. [Crossref] [PubMed]
- Gentner B, Naldini L. Exploiting microRNA regulation for genetic engineering. Tissue Antigens 2012;80:393-403. [Crossref] [PubMed]
- Oulas A, Karathanasis N, Louloupi A, et al. Prediction of miRNA targets. Methods Mol Biol 2015;1269:207-29. [Crossref] [PubMed]
- Melkman-Zehavi T, Oren R, Kredo-Russo S, et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J 2011;30:835-45. [Crossref] [PubMed]
- Karagiannopoulos A, Esguerra JLS, Pedersen MG, et al. Human pancreatic islet miRNA-mRNA networks of altered miRNAs due to glycemic status. iScience 2022;25:103995. [Crossref] [PubMed]
- Wong WKM, Joglekar MV, Saini V, et al. Machine learning workflows identify a microRNA signature of insulin transcription in human tissues. iScience 2021;24:102379. [Crossref] [PubMed]
- Zheng Y, Wang Z, Tu Y, et al. miR-101a and miR-30b contribute to inflammatory cytokine-mediated β-cell dysfunction. Lab Invest 2015;95:1387-97. [Crossref] [PubMed]
- Sebastiani G, Po A, Miele E, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol 2015;52:523-30. [Crossref] [PubMed]
- Yang L, Zhu Y, Kong D, et al. EGF suppresses the expression of miR-124a in pancreatic β cell lines via ETS2 activation through the MEK and PI3K signaling pathways. Int J Biol Sci 2019;15:2561-75. [Crossref] [PubMed]
- Cheung R, Pizza G, Chabosseau P, et al. Glucose-Dependent miR-125b Is a Negative Regulator of β-Cell Function. Diabetes 2022;71:1525-45. [Crossref] [PubMed]
- Tao H, Wang MM, Zhang M, et al. MiR-126 Suppresses the Glucose-Stimulated Proliferation via IRS-2 in INS-1 β Cells. PLoS One 2016;11:e0149954. [Crossref] [PubMed]
- Shen Z, Jiang H, Hsu HT, et al. MicroRNA-127 inhibits cell proliferation via targeting Kif3b in pancreatic β cells. Aging (Albany NY) 2019;11:1342-55. [Crossref] [PubMed]
- Ofori JK, Salunkhe VA, Bagge A, et al. Elevated miR-130a/miR130b/miR-152 expression reduces intracellular ATP levels in the pancreatic beta cell. Sci Rep 2017;7:44986. [Crossref] [PubMed]
- Capuani B, Pacifici F, Della-Morte D, et al. Glucagon Like Peptide 1 and MicroRNA in Metabolic Diseases: Focusing on GLP1 Action on miRNAs. Front Endocrinol (Lausanne) 2018;9:719. [Crossref] [PubMed]
- Mulder NL, Havinga R, Kluiver J, et al. AAV8-mediated gene transfer of microRNA-132 improves beta cell function in mice fed a high-fat diet. J Endocrinol 2019;240:123-32. [Crossref] [PubMed]
- Al-Kafaji G, Al-Muhtaresh HA, Salem AH. Expression and clinical significance of miR-1 and miR-133 in pre-diabetes. Biomed Rep 2021;14:33. [Crossref] [PubMed]
- Fred RG, Bang-Berthelsen CH, Mandrup-Poulsen T, et al. High glucose suppresses human islet insulin biosynthesis by inducing miR-133a leading to decreased polypyrimidine tract binding protein-expression. PLoS One 2010;5:e10843. [Crossref] [PubMed]
- Qiu H, Ma L, Feng F. PICK1 attenuates high glucose-induced pancreatic β-cell death through the PI3K/Akt pathway and is negatively regulated by miR-139-5p. Biochem Biophys Res Commun 2020;522:14-20. [Crossref] [PubMed]
- Yu X, Zhong L. Pioglitazone/microRNA 141/FOXA2: A novel axis in pancreatic β cells proliferation and insulin secretion. Mol Med Rep 2018;17:7931-8. [Crossref] [PubMed]
- Kang MH, Zhang LH, Wijesekara N, et al. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler Thromb Vasc Biol 2013;33:2724-32. [Crossref] [PubMed]
- Lu J, Pang L, Zhang B, et al. Silencing circANKRD36 inhibits streptozotocin-induced insulin resistance and inflammation in diabetic rats by targeting miR-145 via XBP1. Inflamm Res 2021;70:695-704. [Crossref] [PubMed]
- Zou G, Liu T, Guo L, et al. miR-145 modulates lncRNA-ROR and Sox2 expression to maintain human amniotic epithelial stem cell pluripotency and β islet-like cell differentiation efficiency. Gene 2016;591:48-57. [Crossref] [PubMed]
- García-Jacobo RE, Uresti-Rivera EE, Portales-Pérez DP, et al. Circulating miR-146a, miR-34a and miR-375 in type 2 diabetes patients, pre-diabetic and normal-glycaemic individuals in relation to β-cell function, insulin resistance and metabolic parameters. Clin Exp Pharmacol Physiol 2019;46:1092-100. [Crossref] [PubMed]
- Lovis P, Roggli E, Laybutt DR, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 2008;57:2728-36. [Crossref] [PubMed]
- Ruan D, Liu Y, Wang X, et al. miR-149-5p protects against high glucose-induced pancreatic beta cell apoptosis via targeting the BH3-only protein BIM. Exp Mol Pathol 2019;110:104279. [Crossref] [PubMed]
- Sun LL, Jiang BG, Li WT, et al. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res Clin Pract 2011;91:94-100. [Crossref] [PubMed]
- Chen L, Qian H, Xue J, et al. MicroRNA 152 regulates insulin secretion and pancreatic β cell proliferation by targeting PI3Kα. Mol Med Rep 2018;18:4113-21. [Crossref] [PubMed]
- Sun Y, Zhou S, Shi Y, et al. Inhibition of miR-153, an IL-1β-responsive miRNA, prevents beta cell failure and inflammation-associated diabetes. Metabolism 2020;111:154335. [Crossref] [PubMed]
- Xu H, Abuhatzira L, Carmona GN, et al. The Ia-2β intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia 2015;58:2298-306. [Crossref] [PubMed]
- Zhu M, Wei Y, Geißler C, et al. Hyperlipidemia-Induced MicroRNA-155-5p Improves β-Cell Function by Targeting Mafb. Diabetes 2017;66:3072-84. [Crossref] [PubMed]
- Lu Y, Fei XQ, Yang SF, et al. Glucose-induced microRNA-17 promotes pancreatic beta cell proliferation through down-regulation of Menin. Eur Rev Med Pharmacol Sci 2015;19:624-9. [PubMed]
- Chen Y, Tian L, Wan S, et al. MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol Cell Endocrinol 2016;437:213-23. [Crossref] [PubMed]
- Zhang ZW, Zhang LQ, Ding L, et al. MicroRNA-19b downregulates insulin 1 through targeting transcription factor NeuroD1. FEBS Lett 2011;585:2592-8. [Crossref] [PubMed]
- Cheng Y, Hu Q, Zhou J. Silencing of lncRNA PVT1 ameliorates streptozotocin-induced pancreatic β cell injury and enhances insulin secretory capacity by regulating miR-181a-5p. Can J Physiol Pharmacol 2021;99:303-12. [Crossref] [PubMed]
- Li N, Jiang D, He Q, et al. microRNA-181c-5p promotes the formation of insulin-producing cells from human induced pluripotent stem cells by targeting smad7 and TGIF2. Cell Death Dis 2020;11:462. [Crossref] [PubMed]
- Tattikota SG, Rathjen T, Hausser J, et al. miR-184 Regulates Pancreatic β-Cell Function According to Glucose Metabolism. J Biol Chem 2015;290:20284-94. [Crossref] [PubMed]
- Bao L, Fu X, Si M, et al. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One 2015;10:e0116067. [Crossref] [PubMed]
- Locke JM, da Silva Xavier G, Dawe HR, et al. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 2014;57:122-8. [Crossref] [PubMed]
- Li Y, Luo T, Wang L, et al. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic β cells via the inhibition of SOCS3. Int J Mol Med 2016;38:1515-24. [Crossref] [PubMed]
- Wang S, Wei D, Sun X, et al. MiR-190b impedes pancreatic β cell proliferation and insulin secretion by targeting NKX6-1 and may associate to gestational diabetes mellitus. J Recept Signal Transduct Res 2021;41:349-56. [Crossref] [PubMed]
- Panda AC, Sahu I, Kulkarni SD, et al. miR-196b-mediated translation regulation of mouse insulin2 via the 5'UTR. PLoS One 2014;9:e101084. [Crossref] [PubMed]
- Sato-Kunisada R, Yoshida N, Nakamura S, et al. Enhanced Expression of miR-199b-5p Promotes Proliferation of Pancreatic β-Cells by Down-Regulation of MLK3. Microrna 2016;5:57-65. [Crossref] [PubMed]
- Belgardt BF, Ahmed K, Spranger M, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med 2015;21:619-27. [Crossref] [PubMed]
- Ofori JK, Karagiannopoulos A, Nagao M, et al. Human Islet MicroRNA-200c Is Elevated in Type 2 Diabetes and Targets the Transcription Factor ETV5 to Reduce Insulin Secretion. Diabetes 2022;71:275-84. [Crossref] [PubMed]
- Marzinotto I, Pellegrini S, Brigatti C, et al. miR-204 is associated with an endocrine phenotype in human pancreatic islets but does not regulate the insulin mRNA through MAFA. Sci Rep 2017;7:14051. [Crossref] [PubMed]
- Xu G, Chen J, Jing G, et al. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 2013;19:1141-6. [Crossref] [PubMed]
- Ouni M, Gottmann P, Westholm E, et al. MiR-205 is up-regulated in islets of diabetes-susceptible mice and targets the diabetes gene Tcf7l2. Acta Physiol (Oxf) 2021;232:e13693. [Crossref] [PubMed]
- Bai C, Li X, Gao Y, et al. Role of microRNA-21 in the formation of insulin-producing cells from pancreatic progenitor cells. Biochim Biophys Acta 2016;1859:280-93. [Crossref] [PubMed]
- Liu R, Liu C, He X, et al. MicroRNA-21 promotes pancreatic β cell function through modulating glucose uptake. Nat Commun 2022;13:3545. [Crossref] [PubMed]
- Shang J, Li J, Keller MP, et al. Induction of miR-132 and miR-212 Expression by Glucagon-Like Peptide 1 (GLP-1) in Rodent and Human Pancreatic β-Cells. Mol Endocrinol 2015;29:1243-53. [Crossref] [PubMed]
- Zheng H, Li X, Yang X, et al. miR-217/Mafb Axis Involve in High Glucose-Induced β-TC-tet Cell Damage Via Regulating NF-κB Signaling Pathway. Biochem Genet 2020;58:901-13. [Crossref] [PubMed]
- Lang H, Ai Z, You Z, et al. Characterization of miR-218/322-Stxbp1 pathway in the process of insulin secretion. J Mol Endocrinol 2015;54:65-73. [Crossref] [PubMed]
- Fan L, Shan A, Su Y, et al. MiR-221/222 Inhibit Insulin Production of Pancreatic β-Cells in Mice. Endocrinology 2020;161:bqz027. [Crossref] [PubMed]
- Zhao H, Tao S. MiRNA-221 protects islet β cell function in gestational diabetes mellitus by targeting PAK1. Biochem Biophys Res Commun 2019;520:218-24. [Crossref] [PubMed]
- Li Y, Deng S, Peng J, et al. MicroRNA-223 is essential for maintaining functional β-cell mass during diabetes through inhibiting both FOXO1 and SOX6 pathways. J Biol Chem 2019;294:10438-48. [Crossref] [PubMed]
- Grieco FA, Sebastiani G, Juan-Mateu J, et al. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149-5p Regulate the Expression of Proapoptotic BH3-Only Proteins DP5 and PUMA in Human Pancreatic β-Cells. Diabetes 2017;66:100-12. [Crossref] [PubMed]
- Yang J, Lv Y, Zhao Z, et al. A microRNA 24 to secretagogin regulatory pathway mediates cholesterol induced inhibition of insulin secretion. Int J Mol Med 2019;44:608-16. [Crossref] [PubMed]
- Zhu Y, You W, Wang H, et al. MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes 2013;62:3194-206. [Crossref] [PubMed]
- Setyowati Karolina D, Sepramaniam S, Tan HZ, et al. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol 2013;10:1365-78. [Crossref] [PubMed]
- Fu X, Jin L, Wang X, et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci U S A 2013;110:17892-7. [Crossref] [PubMed]
- Yao Y, Xu Y, Wang W, et al. Glucagon-like peptide-1 improves β-cell dysfunction by suppressing the miR-27a-induced downregulation of ATP-binding cassette transporter A1. Biomed Pharmacother 2017;96:497-502. [Crossref] [PubMed]
- Bagge A, Dahmcke CM, Dalgaard LT. Syntaxin-1a is a direct target of miR-29a in insulin-producing β-cells. Horm Metab Res 2013;45:463-6. [Crossref] [PubMed]
- Cheng M, Guo Y, Zhong W, et al. Abnormal Expression of microRNA-296-3p in Type 2 Diabetes Patients and its Role in Pancreatic β-Cells Function by Targeting Tensin Homolog Deleted on Chromosome Ten. Biochem Genet 2022;60:39-53. [Crossref] [PubMed]
- Guo R, Yu Y, Zhang Y, et al. Overexpression of miR-297b-5p protects against stearic acid-induced pancreatic β-cell apoptosis by targeting LATS2. Am J Physiol Endocrinol Metab 2020;318:E430-9. [Crossref] [PubMed]
- Huang Q, You W, Li Y, et al. Glucolipotoxicity-Inhibited miR-299-5p Regulates Pancreatic β-Cell Function and Survival. Diabetes 2018;67:2280-92. [Crossref] [PubMed]
- Kim JW, You YH, Jung S, et al. miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia 2013;56:847-55. [Crossref] [PubMed]
- Mao Y, Schoenborn J, Wang Z, et al. Transgenic overexpression of microRNA-30d in pancreatic beta-cells progressively regulates beta-cell function and identity. Sci Rep 2022;12:11969. [Crossref] [PubMed]
- Tang X, Muniappan L, Tang G, et al. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA 2009;15:287-93. [Crossref] [PubMed]
- Zhao X, Mohan R, Özcan S, et al. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. J Biol Chem 2012;287:31155-64. [Crossref] [PubMed]
- Zou G, Liu T, Guo L, et al. MicroRNA-32 silences WWP2 expression to maintain the pluripotency of human amniotic epithelial stem cells and β islet-like cell differentiation. Int J Mol Med 2018;41:1983-91. [Crossref] [PubMed]
- Li G, Zhang L. miR-335-5p aggravates type 2 diabetes by inhibiting SLC2A4 expression. Biochem Biophys Res Commun 2021;558:71-8. [Crossref] [PubMed]
- Salunkhe VA, Ofori JK, Gandasi NR, et al. MiR-335 overexpression impairs insulin secretion through defective priming of insulin vesicles. Physiol Rep 2017;5:e13493. [Crossref] [PubMed]
- Jacovetti C, Abderrahmani A, Parnaud G, et al. MicroRNAs contribute to compensatory β cell expansion during pregnancy and obesity. J Clin Invest 2012;122:3541-51. [Crossref] [PubMed]
- Jacovetti C, Jimenez V, Ayuso E, et al. Contribution of Intronic miR-338-3p and Its Hosting Gene AATK to Compensatory β-Cell Mass Expansion. Mol Endocrinol 2015;29:693-702. [Crossref] [PubMed]
- Kong X, Liu CX, Wang GD, et al. LncRNA LEGLTBC Functions as a ceRNA to Antagonize the Effects of miR-34a on the Downregulation of SIRT1 in Glucolipotoxicity-Induced INS-1 Beta Cell Oxidative Stress and Apoptosis. Oxid Med Cell Longev 2019;2019:4010764. [Crossref] [PubMed]
- Sun X, Ji G, Li P, et al. miR-344-5p Modulates Cholesterol-Induced β-Cell Apoptosis and Dysfunction Through Regulating Caveolin-1 Expression. Front Endocrinol (Lausanne) 2021;12:695164. [Crossref] [PubMed]
- El Ouaamari A, Baroukh N, Martens GA, et al. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008;57:2708-17. [Crossref] [PubMed]
- Li Y, Xu X, Liang Y, et al. miR-375 enhances palmitate-induced lipoapoptosis in insulin-secreting NIT-1 cells by repressing myotrophin (V1) protein expression. Int J Clin Exp Pathol 2010;3:254-64. [PubMed]
- Nathan G, Kredo-Russo S, Geiger T, et al. MiR-375 promotes redifferentiation of adult human β cells expanded in vitro. PLoS One 2015;10:e0122108. [Crossref] [PubMed]
- Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226-30. [Crossref] [PubMed]
- Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A 2009;106:5813-8. [Crossref] [PubMed]
- Sedgeman LR, Beysen C, Ramirez Solano MA, et al. Beta cell secretion of miR-375 to HDL is inversely associated with insulin secretion. Sci Rep 2019;9:3803. [Crossref] [PubMed]
- Zhang ZW, Men T, Feng RC, et al. miR-375 inhibits proliferation of mouse pancreatic progenitor cells by targeting YAP1. Cell Physiol Biochem 2013;32:1808-17. [Crossref] [PubMed]
- Zhao H, Guan J, Lee HM, et al. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through beta-cell deficit and islet amyloid deposition. Pancreas 2010;39:843-6. [Crossref] [PubMed]
- Cheng X, Huang Y, Yang P, et al. miR-383 ameliorates high glucose-induced β-cells apoptosis and hyperglycemia in high-fat induced diabetic mice. Life Sci 2020;263:118571. [Crossref] [PubMed]
- Sun R, Xue W, Zhao J. Hsa_circ_0054633 mediates apoptosis and insulin secretion in human pancreatic β cells through miR-409-3p/caspase-8 axis. Diabetes Res Clin Pract 2021;176:108837. [Crossref] [PubMed]
- Wang M. miR-433 protects pancreatic β cell growth in high-glucose conditions. Mol Med Rep 2017;16:2604-10. [Crossref] [PubMed]
- Hu Q, Mu J, Liu Y, et al. Obesity-Induced miR-455 Upregulation Promotes Adaptive Pancreatic β-Cell Proliferation Through the CPEB1/CDKN1B Pathway. Diabetes 2022;71:394-411. [Crossref] [PubMed]
- Hou X, Wu W, Yin B, et al. MicroRNA-463-3p/ABCG4: A new axis in glucose-stimulated insulin secretion. Obesity (Silver Spring) 2016;24:2368-76. [Crossref] [PubMed]
- Mohan R, Mao Y, Zhang S, et al. Differentially Expressed MicroRNA-483 Confers Distinct Functions in Pancreatic β- and α-Cells. J Biol Chem 2015;290:19955-66. [Crossref] [PubMed]
- Wang Z, Mohan R, Chen X, et al. microRNA-483 Protects Pancreatic β-Cells by Targeting ALDH1A3. Endocrinology 2021;162:bqab031. [Crossref] [PubMed]
- Zhong Z, Su W, Chen H. MicroRNA 532 5p regulates oxidative stress and insulin secretion damage in high glucose induced pancreatic β cells by downregulating the expression levels of CCND1. Mol Med Rep 2021;24:793. [Crossref] [PubMed]
- Ren L. Circular RNA PIP5K1A act as microRNA-552-3p sponge to regulates inflammation, oxidative damage in glucolipotoxicity-induced pancreatic INS-1 β-cells via Janus kinase 1. Bioengineered 2022;13:5724-36. [Crossref] [PubMed]
- Chen XY, Li GM, Dong Q, et al. MiR-577 inhibits pancreatic β-cell function and survival by targeting fibroblast growth factor 21 (FGF-21) in pediatric diabetes. Genet Mol Res 2015;14:15462-70. [Crossref] [PubMed]
- Shaer A, Azarpira N, Karimi MH, et al. Differentiation of Human-Induced Pluripotent Stem Cells Into Insulin-Producing Clusters by MicroRNA-7. Exp Clin Transplant 2016;14:555-63. [PubMed]
- Latreille M, Hausser J, Stützer I, et al. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest 2014;124:2722-35. [Crossref] [PubMed]
- Wang Y, Liu J, Liu C, et al. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes 2013;62:887-95. [Crossref] [PubMed]
- Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. [Crossref] [PubMed]
- Rodríguez-Comas J, Moreno-Asso A, Moreno-Vedia J, et al. Stress-Induced MicroRNA-708 Impairs β-Cell Function and Growth. Diabetes 2017;66:3029-40. [Crossref] [PubMed]
- Zhang F, Ma D, Zhao W, et al. Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat Commun 2020;11:1822. [Crossref] [PubMed]
- Hu D, Wang Y, Zhang H, et al. Identification of miR-9 as a negative factor of insulin secretion from beta cells. J Physiol Biochem 2018;74:291-9. [Crossref] [PubMed]
- Plaisance V, Abderrahmani A, Perret-Menoud V, et al. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem 2006;281:26932-42. [Crossref] [PubMed]
- Wang W, Wang J, Yan M, et al. MiRNA-92a protects pancreatic B-cell function by targeting KLF2 in diabetes mellitus. Biochem Biophys Res Commun 2018;500:577-82. [Crossref] [PubMed]
- Qi H, Yao L, Liu Q. MicroRNA-96 regulates pancreatic β cell function under the pathological condition of diabetes mellitus through targeting Foxo1 and Sox6. Biochem Biophys Res Commun 2019;519:294-301. [Crossref] [PubMed]
- Aljaibeji H, Elemam NM, Mohammed AK, et al. Let7b-5p is Upregulated in the Serum of Emirati Patients with Type 2 Diabetes and Regulates Insulin Secretion in INS-1 Cells. Exp Clin Endocrinol Diabetes 2022;130:22-9. [Crossref] [PubMed]
- Ji H, Fan L, Shan A, et al. Let7b-5p inhibits insulin secretion and decreases pancreatic β-cell mass in mice. Mol Cell Endocrinol 2022;540:111506. [Crossref] [PubMed]
- Frampton AE, Krell J, Jamieson NB, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer 2015;51:1389-404. [Crossref] [PubMed]
- Sempere LF, Powell K, Rana J, et al. Role of non-coding RNAs in tumor progression and metastasis in pancreatic cancer. Cancer Metastasis Rev 2021;40:761-76. [Crossref] [PubMed]
- Taucher V, Mangge H, Haybaeck J. Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell Oncol (Dordr) 2016;39:295-318. [Crossref] [PubMed]
- Lu H, Guo R, Zhang Y, et al. Inhibition of lncRNA TCONS_00077866 Ameliorates the High Stearic Acid Diet-Induced Mouse Pancreatic β-Cell Inflammatory Response by Increasing miR-297b-5p to Downregulate SAA3 Expression. Diabetes 2021;70:2275-88. [Crossref] [PubMed]
- Erener S, Mojibian M, Fox JK, et al. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology 2013;154:603-8. [Crossref] [PubMed]
- Haniff HS, Liu X, Tong Y, et al. A structure-specific small molecule inhibits a miRNA-200 family member precursor and reverses a type 2 diabetes phenotype. Cell Chem Biol 2022;29:300-311.e10. [Crossref] [PubMed]
- Tang XW, Qin QX. miR-335-5p induces insulin resistance and pancreatic islet β-cell secretion in gestational diabetes mellitus mice through VASH1-mediated TGF-β signaling pathway. J Cell Physiol 2019;234:6654-66. [Crossref] [PubMed]
- Zhang YL, Chen XQ. Dysregulation of microRNA-770-5p influences pancreatic-β-cell function by targeting TP53 regulated inhibitor of apoptosis 1 in gestational diabetes mellitus. Eur Rev Med Pharmacol Sci 2020;24:793-801. [PubMed]
- Aguayo-Mazzucato C, Bonner-Weir S. Pancreatic β Cell Regeneration as a Possible Therapy for Diabetes. Cell Metab 2018;27:57-67. [Crossref] [PubMed]
- Nihad M, Shenoy P S, Bose B. Cell therapy research for Diabetes: Pancreatic β cell differentiation from pluripotent stem cells. Diabetes Res Clin Pract 2021;181:109084. [Crossref] [PubMed]
- Vakilian M, Tahamtani Y, Ghaedi K. A review on insulin trafficking and exocytosis. Gene 2019;706:52-61. [Crossref] [PubMed]
- Malm HA, Mollet IG, Berggreen C, et al. Transcriptional regulation of the miR-212/miR-132 cluster in insulin-secreting β-cells by cAMP-regulated transcriptional co-activator 1 and salt-inducible kinases. Mol Cell Endocrinol 2016;424:23-33. [Crossref] [PubMed]
- Cao M, Mao Z, Kam C, et al. PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol 2013;11:e1001541. [Crossref] [PubMed]
- Esguerra JL, Bolmeson C, Cilio CM, et al. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 2011;6:e18613. [Crossref] [PubMed]
- Downing S, Zhang F, Chen Z, et al. MicroRNA-7 directly targets Reg1 in pancreatic cells. Am J Physiol Cell Physiol 2019;317:C366-74. [Crossref] [PubMed]
- Brunham LR, Kruit JK, Pape TD, et al. Beta-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med 2007;13:340-7. [Crossref] [PubMed]
- Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet 2014;383:69-82. [Crossref] [PubMed]
- Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol 2009;5:219-26. [Crossref] [PubMed]
- Mirzaei R, Zamani F, Hajibaba M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J Neuroimmunol 2021;358:577640. [Crossref] [PubMed]
- Azhir Z, Dehghanian F, Hojati Z. Increased expression of microRNAs, miR-20a and miR-326 in PBMCs of patients with type 1 diabetes. Mol Biol Rep 2018;45:1973-80. [Crossref] [PubMed]
- Sebastiani G, Grieco FA, Spagnuolo I, et al. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev 2011;27:862-6. [Crossref] [PubMed]
- Fornari TA, Donate PB, Assis AF, et al. Comprehensive Survey of miRNA-mRNA Interactions Reveals That Ccr7 and Cd247 (CD3 zeta) are Posttranscriptionally Controlled in Pancreas Infiltrating T Lymphocytes of Non-Obese Diabetic (NOD) Mice. PLoS One 2015;10:e0142688. [Crossref] [PubMed]
- Scherm MG, Serr I, Zahm AM, et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat Commun 2019;10:5697. [Crossref] [PubMed]
- Wang P, Liu Q, Zhao H, et al. miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci Rep 2020;10:5302. [Crossref] [PubMed]
- Ruan Q, Wang T, Kameswaran V, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci U S A 2011;108:12030-5. [Crossref] [PubMed]
- Duan X, Zhao L, Jin W, et al. MicroRNA-203a regulates pancreatic β cell proliferation and apoptosis by targeting IRS2. Mol Biol Rep 2020;47:7557-66. [Crossref] [PubMed]
- Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008;9:405-14. [Crossref] [PubMed]
- Serr I, Fürst RW, Ott VB, et al. miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity. Proc Natl Acad Sci U S A 2016;113:E6659-68. [Crossref] [PubMed]
- Guay C, Kruit JK, Rome S, et al. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic β Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab 2019;29:348-361.e6. [Crossref] [PubMed]
- Gao X, Zhao S. miRNA-16-5p inhibits the apoptosis of high glucose-induced pancreatic β cells via targeting of CXCL10: potential biomarkers in type 1 diabetes mellitus. Endokrynol Pol 2020;71:404-10. [Crossref] [PubMed]
- Tian J, Pan W, Xu X, et al. NF-κB inhibits the occurrence of type 1 diabetes through microRNA-150-dependent PUMA degradation. Life Sci 2020;255:117724. [Crossref] [PubMed]
- Yu CY, Yang CY, Rui ZL. MicroRNA-125b-5p improves pancreatic β-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci 2019;224:67-75. [Crossref] [PubMed]
- Ventriglia G, Mancarella F, Sebastiani G, et al. miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2020;63:124-36. [Crossref] [PubMed]
- Bagge A, Clausen TR, Larsen S, et al. MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochem Biophys Res Commun 2012;426:266-72. [Crossref] [PubMed]
- Roggli E, Gattesco S, Caille D, et al. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes 2012;61:1742-51. [Crossref] [PubMed]
- Dooley J, Garcia-Perez JE, Sreenivasan J, et al. The microRNA-29 Family Dictates the Balance Between Homeostatic and Pathological Glucose Handling in Diabetes and Obesity. Diabetes 2016;65:53-61. [Crossref] [PubMed]
- Pandey AK, Verma G, Vig S, et al. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol Cell Endocrinol 2011;332:125-33. [Crossref] [PubMed]
- Ma H, Zhang S, Shi D, et al. MicroRNA-26a Promotes Regulatory T cells and Suppresses Autoimmune Diabetes in Mice. Inflammation 2016;39:1-9. [Crossref] [PubMed]
- Song Y, Jin D, Jiang X, et al. Overexpression of microRNA-26a protects against deficient β-cell function via targeting phosphatase with tensin homology in mouse models of type 2 diabetes. Biochem Biophys Res Commun 2018;495:1312-6. [Crossref] [PubMed]
- Xu H, Du X, Xu J, et al. Pancreatic β cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving β cell function. PLoS Biol 2020;18:e3000603. [Crossref] [PubMed]
- Gál E, Dolenšek J, Stožer A, et al. Mechanisms of Post-Pancreatitis Diabetes Mellitus and Cystic Fibrosis-Related Diabetes: A Review of Preclinical Studies. Front Endocrinol (Lausanne) 2021;12:715043. [Crossref] [PubMed]
- Lankisch PG, Apte M, Banks PA. Acute pancreatitis. Lancet 2015;386:85-96. [Crossref] [PubMed]
- Beyer G, Habtezion A, Werner J, et al. Chronic pancreatitis. Lancet 2020;396:499-512. [Crossref] [PubMed]
- Du W, Liu G, Shi N, et al. A microRNA checkpoint for Ca2+ signaling and overload in acute pancreatitis. Mol Ther 2022;30:1754-74. [Crossref] [PubMed]
- Dai J, Jiang M, Hu Y, et al. Dysregulated SREBP1c/miR-153 signaling induced by hypertriglyceridemia worsens acute pancreatitis and delays tissue repair. JCI Insight 2021;6:138584. [Crossref] [PubMed]
- Wu D, Guo J, Qi B, et al. TGF-β1 induced proliferation, migration, and ECM accumulation through the SNHG11/miR-34b/LIF pathway in human pancreatic stellate cells. Endocr J 2021;68:1347-57. [Crossref] [PubMed]
- Zhu X, Liu D, Li G, et al. Exosomal miR-140-3p and miR-143-3p from TGF-β1-treated pancreatic stellate cells target BCL2 mRNA to increase β-cell apoptosis. Mol Cell Endocrinol 2022;551:111653. [Crossref] [PubMed]
- Chidester S, Livinski AA, Fish AF, et al. The Role of Extracellular Vesicles in β-Cell Function and Viability: A Scoping Review. Front Endocrinol (Lausanne) 2020;11:375. [Crossref] [PubMed]
- Wu Y, Huang Q, Bu S. Cross talk between exosomes and pancreatic β-cells in diabetes. Arch Physiol Biochem 2022;128:1140-9. [Crossref] [PubMed]
- Guay C, Menoud V, Rome S, et al. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun Signal 2015;13:17. [Crossref] [PubMed]
- Sun Y, Zhou Y, Shi Y, et al. Expression of miRNA-29 in Pancreatic β Cells Promotes Inflammation and Diabetes via TRAF3. Cell Rep 2021;34:108576. [Crossref] [PubMed]
- Li J, Zhang Y, Ye Y, et al. Pancreatic β cells control glucose homeostasis via the secretion of exosomal miR-29 family. J Extracell Vesicles 2021;10:e12055. [Crossref] [PubMed]
- Fu X, Dong B, Tian Y, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest 2015;125:2497-509. [Crossref] [PubMed]
- Zhang A, Li D, Liu Y, et al. Islet β cell: An endocrine cell secreting miRNAs. Biochem Biophys Res Commun 2018;495:1648-54. [Crossref] [PubMed]
- Jalabert A, Vial G, Guay C, et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 2016;59:1049-58. [Crossref] [PubMed]
- Lau J, Kawahira H, Hebrok M. Hedgehog signaling in pancreas development and disease. Cell Mol Life Sci 2006;63:642-52. [Crossref] [PubMed]
- Spinetti G, Fortunato O, Caporali A, et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res 2013;112:335-46. [Crossref] [PubMed]
- Kamalden TA, Macgregor-Das AM, Kannan SM, et al. Exosomal MicroRNA-15a Transfer from the Pancreas Augments Diabetic Complications by Inducing Oxidative Stress. Antioxid Redox Signal 2017;27:913-30. [Crossref] [PubMed]
- Gesmundo I, Pardini B, Gargantini E, et al. Adipocyte-derived extracellular vesicles regulate survival and function of pancreatic β cells. JCI Insight 2021;6:141962. [Crossref] [PubMed]
- Kulkarni RN, Mizrachi EB, Ocana AG, et al. Human β-cell proliferation and intracellular signaling: driving in the dark without a road map. Diabetes 2012;61:2205-13. [Crossref] [PubMed]
- Qian B, Yang Y, Tang N, et al. M1 macrophage-derived exosomes impair beta cell insulin secretion via miR-212-5p by targeting SIRT2 and inhibiting Akt/GSK-3β/β-catenin pathway in mice. Diabetologia 2021;64:2037-51. [Crossref] [PubMed]
- Gao H, Luo Z, Jin Z, et al. Adipose Tissue Macrophages Modulate Obesity-Associated β Cell Adaptations through Secreted miRNA-Containing Extracellular Vesicles. Cells 2021;10:2451. [Crossref] [PubMed]
- Tsukita S, Yamada T, Takahashi K, et al. MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic β-Cell Proliferation. EBioMedicine 2017;15:163-72. [Crossref] [PubMed]
- Kitaguchi T, Oya M, Wada Y, et al. Extracellular calcium influx activates adenylate cyclase 1 and potentiates insulin secretion in MIN6 cells. Biochem J 2013;450:365-73. [Crossref] [PubMed]
- Seino S, Shibasaki T. PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol Rev 2005;85:1303-42. [Crossref] [PubMed]
- Pang W, Yao W, Dai X, et al. Pancreatic cancer-derived exosomal microRNA-19a induces β-cell dysfunction by targeting ADCY1 and EPAC2. Int J Biol Sci 2021;17:3622-33. [Crossref] [PubMed]
- Su J, Pang W, Zhang A, et al. Exosomal miR-19a decreases insulin production by targeting Neurod1 in pancreatic cancer associated diabetes. Mol Biol Rep 2022;49:1711-20. [Crossref] [PubMed]
- Cao M, Isaac R, Yan W, et al. Cancer-cell-secreted extracellular vesicles suppress insulin secretion through miR-122 to impair systemic glucose homeostasis and contribute to tumour growth. Nat Cell Biol 2022;24:954-67. [Crossref] [PubMed]
- Chen L, Heikkinen L, Wang C, et al. Trends in the development of miRNA bioinformatics tools. Brief Bioinform 2019;20:1836-52. [Crossref] [PubMed]
- Pal AS, Kasinski AL. Animal Models to Study MicroRNA Function. Adv Cancer Res 2017;135:53-118. [Crossref] [PubMed]
- Parveen A, Mustafa SH, Yadav P, et al. Applications of Machine Learning in miRNA Discovery and Target Prediction. Curr Genomics 2019;20:537-44. [Crossref] [PubMed]
- Nair G, Hebrok M. Islet formation in mice and men: lessons for the generation of functional insulin-producing β-cells from human pluripotent stem cells. Curr Opin Genet Dev 2015;32:171-80. [Crossref] [PubMed]
- Adams MT, Blum B. Determinants and dynamics of pancreatic islet architecture. Islets 2022;14:82-100. [Crossref] [PubMed]
- Arrojo e Drigo R, Ali Y, Diez J, et al. New insights into the architecture of the islet of Langerhans: a focused cross-species assessment. Diabetologia 2015;58:2218-28. [Crossref] [PubMed]
- Dolenšek J, Rupnik MS, Stožer A. Structural similarities and differences between the human and the mouse pancreas. Islets 2015;7:e1024405. [Crossref] [PubMed]
- Tarifeño-Saldivia E, Lavergne A, Bernard A, et al. Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes. BMC Biol 2017;15:21. [Crossref] [PubMed]
- Eizirik DL, Pipeleers DG, Ling Z, et al. Major species differences between humans and rodents in the susceptibility to pancreatic beta-cell injury. Proc Natl Acad Sci U S A 1994;91:9253-6. [Crossref] [PubMed]
- Hunter CS, Stein RW. Evidence for Loss in Identity, De-Differentiation, and Trans-Differentiation of Islet β-Cells in Type 2 Diabetes. Front Genet 2017;8:35. [Crossref] [PubMed]
- Moin ASM, Butler AE. Alterations in Beta Cell Identity in Type 1 and Type 2 Diabetes. Curr Diab Rep 2019;19:83. [Crossref] [PubMed]
- Keller DM, Perez IG. Dual regulation of miR-375 and CREM genes in pancreatic beta cells. Islets 2022;14:139-48. [Crossref] [PubMed]
- Mir MM, Mir R, Alghamdi MAA, et al. Potential impact of GCK, MIR-196A-2 and MIR-423 gene abnormalities on the development and progression of type 2 diabetes mellitus in Asir and Tabuk regions of Saudi Arabia. Mol Med Rep 2022;25:162. [Crossref] [PubMed]
- de Candia P, Spinetti G, Specchia C, et al. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS One 2017;12:e0188980. [Crossref] [PubMed]
- Grieco GE, Cataldo D, Ceccarelli E, et al. Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA 2018;4:37. [Crossref] [PubMed]
- Melnik BC. Milk exosomal miRNAs: potential drivers of AMPK-to-mTORC1 switching in β-cell de-differentiation of type 2 diabetes mellitus. Nutr Metab (Lond) 2019;16:85. [Crossref] [PubMed]
- Zhang L, Chen T, Yin Y, et al. Dietary microRNA-A Novel Functional Component of Food. Adv Nutr 2019;10:711-21. [Crossref] [PubMed]
- Zhang S, Cheng Z, Wang Y, et al. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des Devel Ther 2021;15:721-33. [Crossref] [PubMed]
Cite this article as: Du RQ, Vidal-Puig A, Rodriguez-Cuenca S. miRNAs provide mechanisms for integrated control of endocrine pancreas homeostasis and metabolic disease pathogenesis: a narrative review. ExRNA 2022;4:24.


